The data in a Gen3 data commons can be browsed and downloaded using several different methods. The following general documentation will cover some standard methods of data access in a Gen3 data commons. Ultimately, however, the methods of data access offered in a Gen3 data commons is determined by agreements made between the data commons’ sponsors and data contributors.
Various levels of data access can be configured in a Gen3 data commons using the Fence and Arborist services. If open access data is hosted, a data commons can be configured to allow anonymous access to data, which means users can explore data without logging in. This is the case for the Open Access Data Commons .
In cases where data is controlled access, typically external users will receive instructions on how to access data and may be required to sign a DUA (Data Use Agreement) legal document.
The following sections provide details on how to explore and access data from within the data commons website and from the command-line by sending requests to the Gen3 open APIs.
The Gen3 software stack offers a data portal service that creates a website with graphical tools for performing the basic functionality of a data commons, like browsing data in projects, building patient cohorts across projects, downloading metadata or data files for cohorts, and building database queries.
The primary tool for exploring data within a Gen3 data commons is the Exploration Page, which offers faceted search of data across projects, for example, https://gen3.datacommons.io/explorer . This page can be accessed from the /explorer endpoint or the top navigation bar, by clicking on the “Exploration” icon.

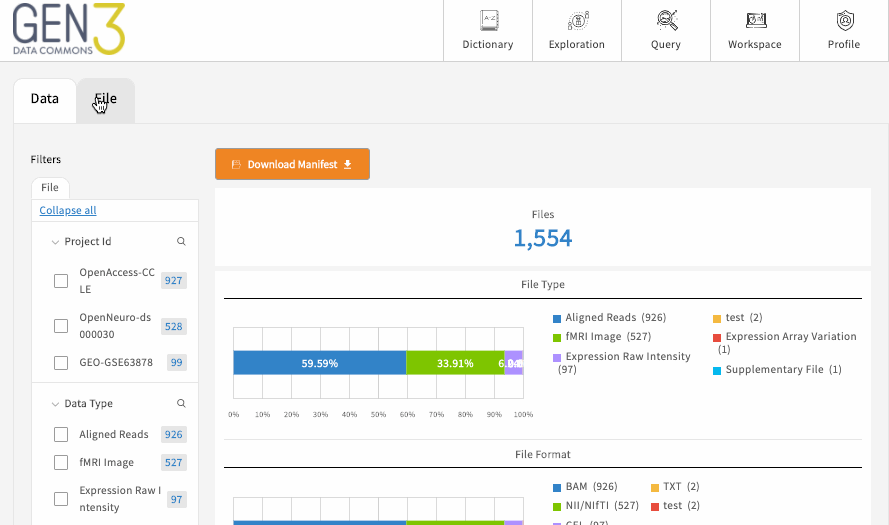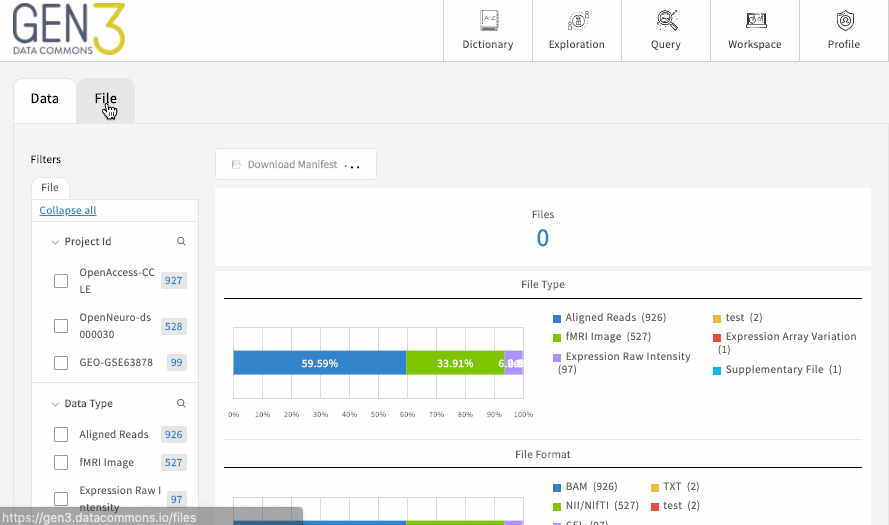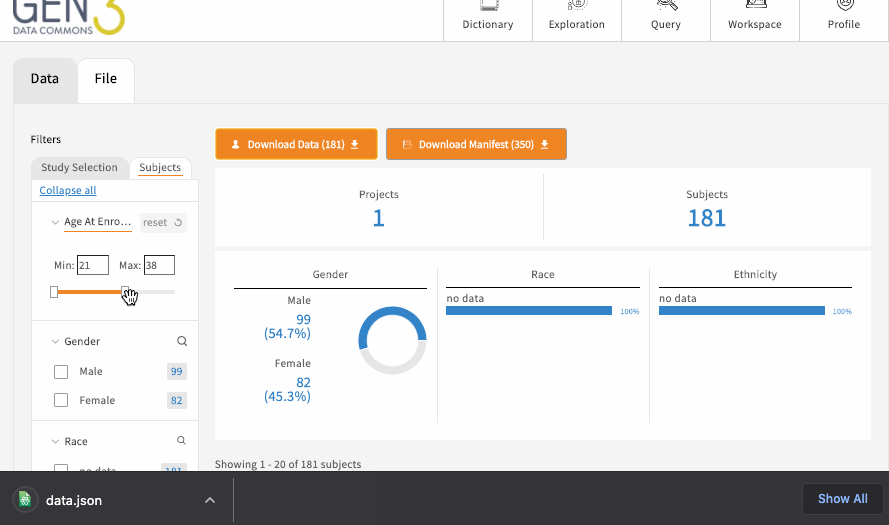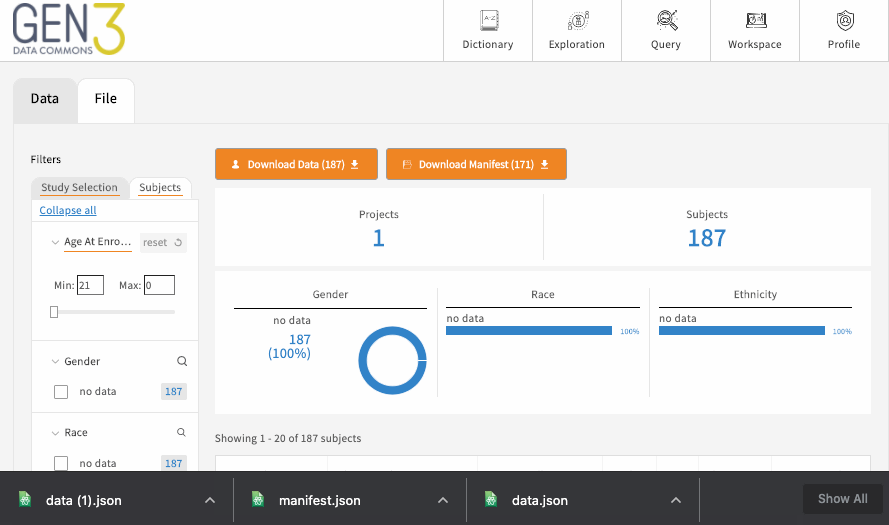
The exploration page has one or several tabs at the top, which each represent a flattened ElasticSearch document of structured metadata records across all the projects in the data commons, which is displayed as a table towards the bottom center of the page. For example, there may be a “Subjects” tab for building patient cohorts, which displays a table of all the records and associated metadata in the subject node, like medical history and demographics. Most commons also have one or more “File” tabs for filtering all the files in a data commons based on things like the file format, data type, or other linked contextual variables, like linked patient demographics or medical history.
Each of these main tabs will have filters on the left-hand side, which can be used to filter the data displayed in the table. There may be an optional button on each tab to download the flattened metadata table as a JSON file. This button should download the table in its filtered state. To remove a filter, click “clear” on individual facets, and you can remove all filters by reloading the page.
Note: The main tabs in the Exploration Page, the available filters, and the properties listed in the data table are entirely customizable and will be different for various Gen3 data commons.
If the table is a list of files, there should be a button for downloading a JSON file that serves as a manifest to use with the gen3-client for downloading multiple files . Otherwise, to download single a file listed in the table, simply click on the GUID (globally unique identifier, or object_id), which should open a page with a download button.
Note: Some data commons have security measures in place that limit what environments users can access data files. For example, users may be required to download and analyze data files in a protected environment, such as a virtual machines (VM) in a virtual private cloud (VPC) or in the built-in Gen3 Workspace, which is accessed by clicking on “Workspace” in the top navigation bar of the data commons website. For more information on the Workspace, see the documentation on how to access and use the Gen3 Workspace .
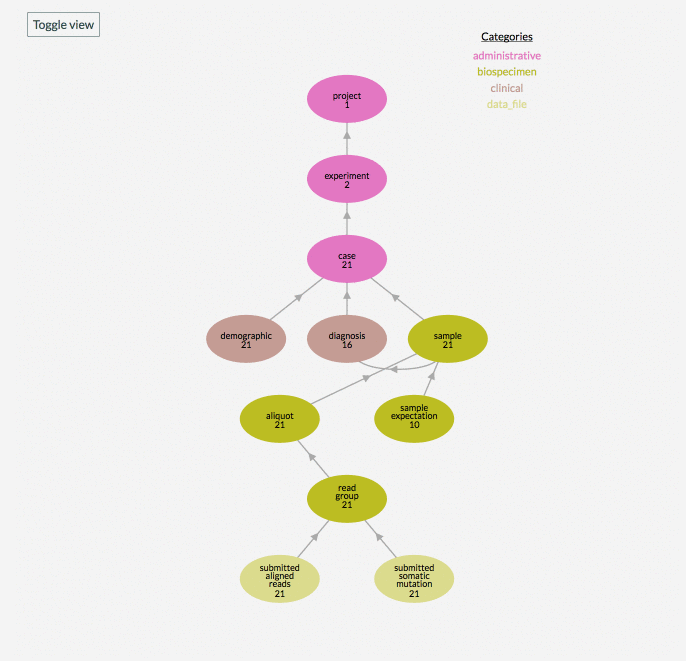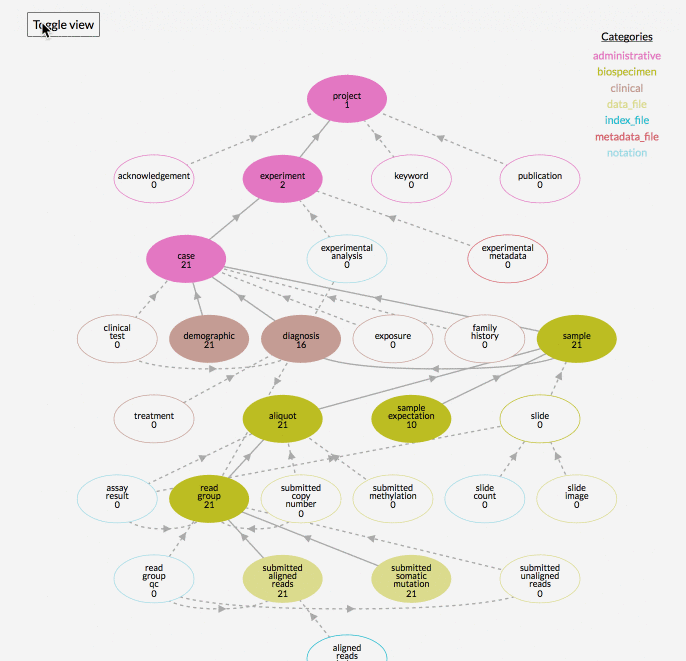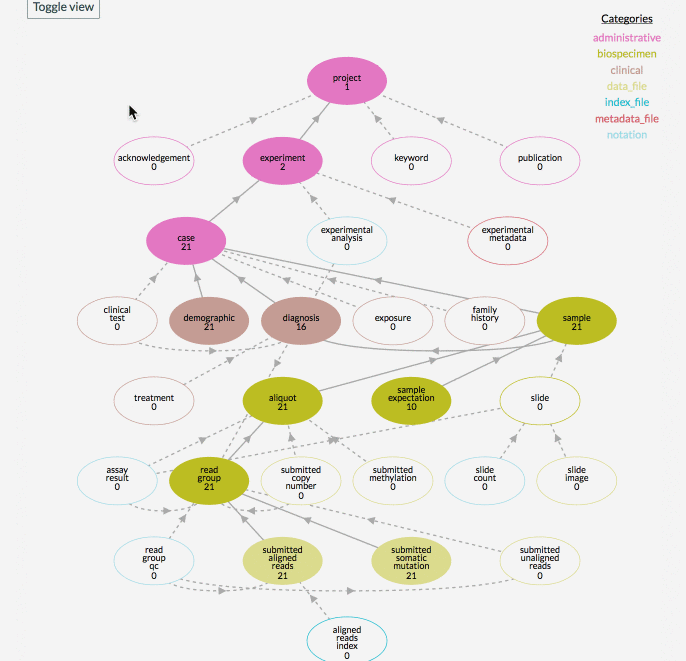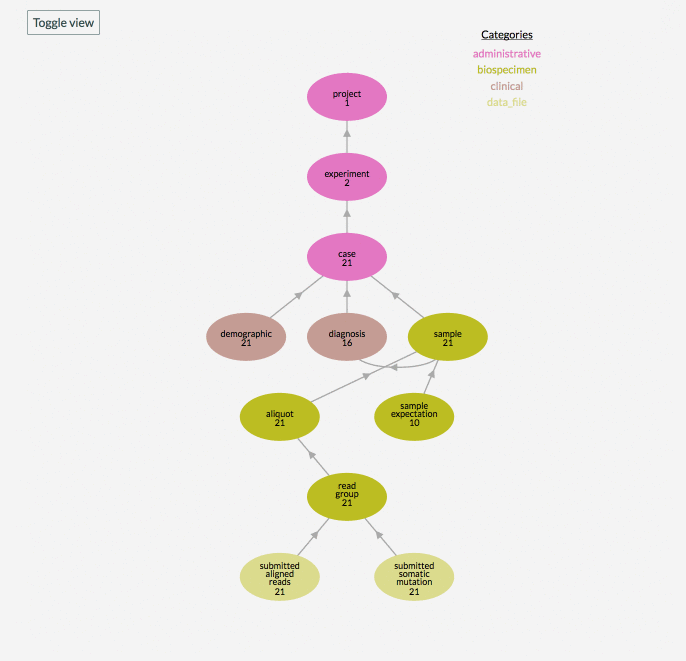
A graphical model of the structured data in individual data projects of a data commons can be browsed by navigating to the /submission endpoint of a data commons website or by clicking on the “Browse Data” or “Submit Data” buttons in the top navigation bar, for example, https://gen3.datacommons.io/submission . This page lists all the data projects within a commons a user has authorization to view. Clicking the “Browse” or “Submit Data” button by a project ID will open a view of that individual project’s structured metadata graph, which can be further inspected by clicking on a node in the graph model and then viewing individual records by clicking “View” by the submitter_id or downloading all the records in that node by clicking the “Download All” button.
Note: Users who are authorized to submit data may see a “Submit Data” button instead of “Browse Data”, and will also be able to upload or create structured data in the project on this page.
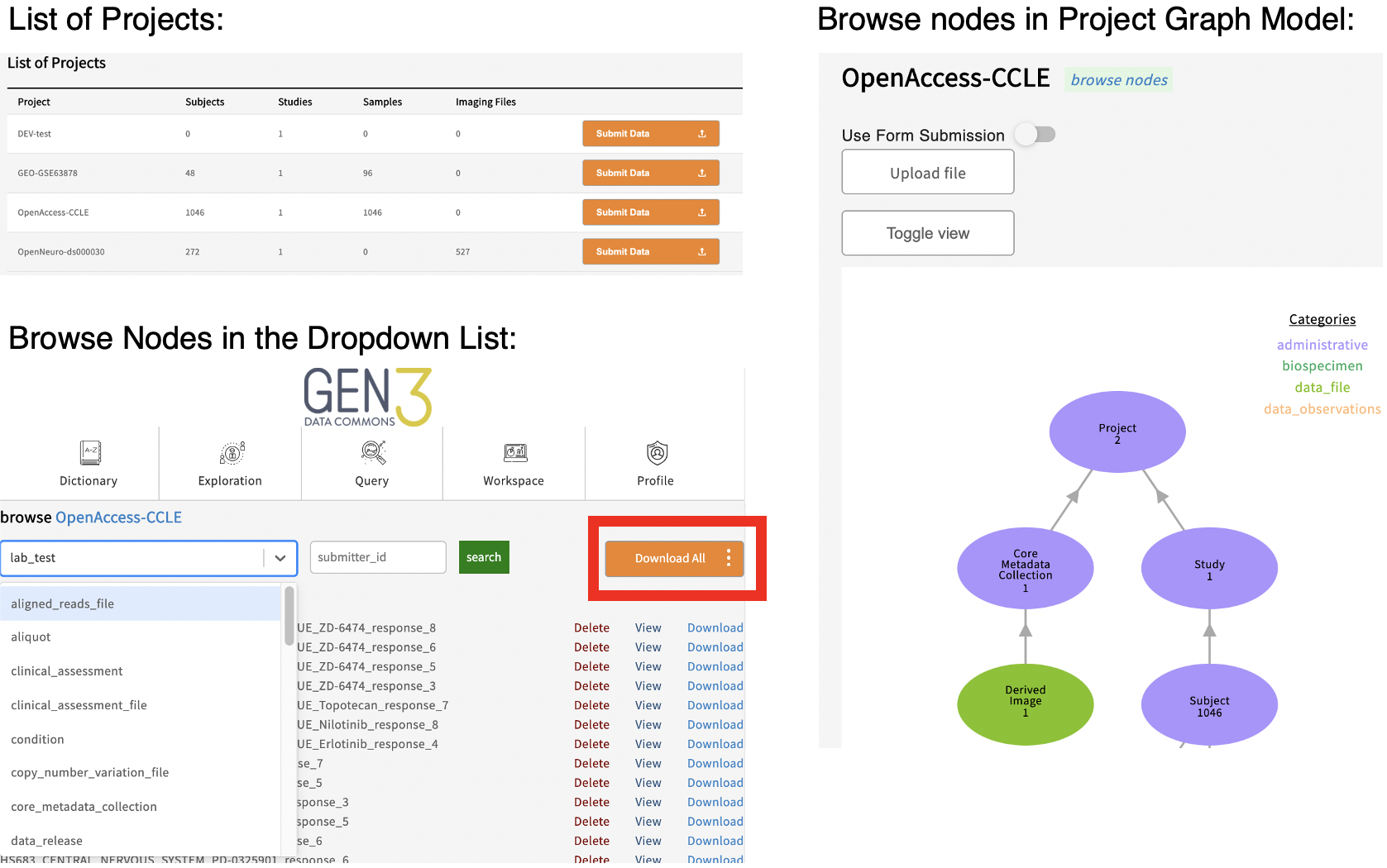
In the graphical model of a data project, the number you see underneath the node name, for example ‘subject’, reflects the number of records in that node of the project. The “Toggle View” button is used to show or hide nodes in the data model that the project has no records for.
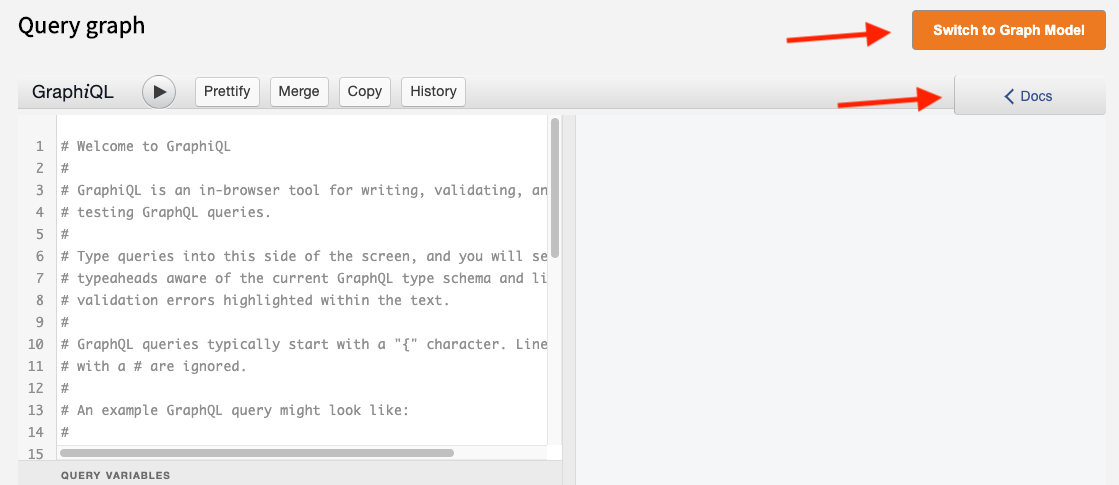
The structured data in a Gen3 data commons can be queried by using the graphQL query language within the GraphiQL interface for building queries, which can be accessed by clicking the “Query” button in the top navigation bar or by navigating to the /query endpoint, for example, https://gen3.datacommons.io/query .

The button on the Query page to switch between the “Graph Model” or “Flat Model”, will direct the queries to different databases, namely Postgres and ElasticSearch, respectively. Pressing the “Docs” button will reveal documentation of the data commons graphQL schema, which will list the queryable nodes and properties.
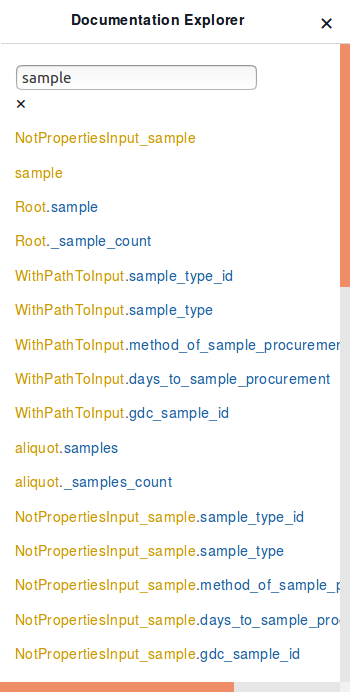
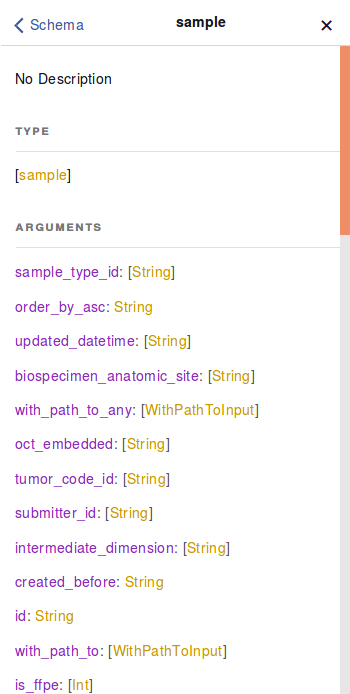
For example, typing the name of a node, “sample”, into the “Search Schema” search-box in the Graph Model, and clicking the “root.sample” option will display all the properties that can be queried for that node.
The following example query returns the subject and submitter ids in the subject node in the data commons:
{
subject {
subject_id
submitter_id
}
}
More detailed information on how to query specific data can be found here .
The beauty of a Gen3 data commons is that all the functionality of the data commons website is available by sending requests to the open APIs of the data commons. Detailed API specifications of the Gen3 services can be browsed in the developer documentation .
Users can submit queries to the Gen3 APIs to access structured data across the projects in the data commons. Queries can be sent using, for example, the Python “requests” package or similar functions in other programming languages. Further details on how to send queries to the API are documented here .
Users with “read” access to a project can export entire structured metadata records by sending requests to the API. Single records can be exported or all records in a specific node of a project can be retrieved. For more information, see the documentation on using the API .
To make sending requests to the Gen3 APIs easier, the bioinformatics team at the Center for Translational Data Science (CTDS) at University of Chicago has put together a basic Python SDK (software development kit) to help users interact with the Gen3 APIs.
The SDK is essentially a collection of Python wrapper functions for sending requests to the API. It is open source and can be found on Github . Thorough documentation for the SDK can be found in the GitHub repository documentation page .
The Gen3Submission class of the Python SDK has functions for sending queries to the API and also for retrieving the graphQL schema of the data commons. Queries can be used to pinpoint specific data of interest by providing query arguments that act as filters on records in the database and providing lists of properties to retrieve for those records. If all the structured data in a record or node is desired, as opposed to only specific properties, then see the export functions below.
Entire structured data records can be exported as a JSON or TSV file using the
Gen3Submission Python SDK class
. The
export_record
function will export a single structured metadata record in a specific node of a specific project, whereas the
export_node
function will export all the structured metadata records in a specified node of a specific project.
More SDK examples and how to get started with the SDK can be also found in the analyze-data section .