The easiest way to query metadata in a Gen3 data commons is done by using the graphQL query language with the GraphiQL interface, which can be accessed by clicking “Query” in the top navigation bar or by navigating to the URL: https://gen3.datacommons.io/query . The URL https://gen3.datacommons.io can be replaced with the URL of other Gen3 data commons.
This query portal has been optimized to autocomplete fields based on content, increase speed and responsiveness, pass variables, and generally make it easier for users to find information. The “Docs” button will display documentation of the queryable nodes and properties. From the GraphiQL interface of the data portal, you can switch between Graph Model or Flat Model –each using endpoints that query different databases (Postgres and ElasticSearch, respectively). Notably, the same queries can be sent to both the flat and graph model API endpoints from the command-line.
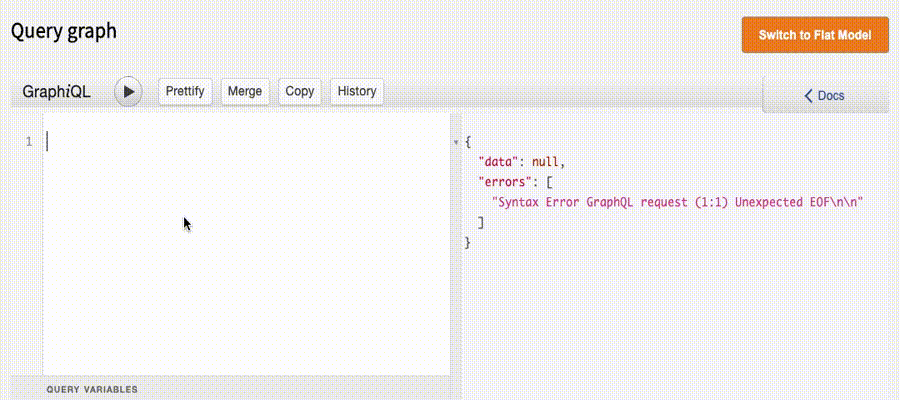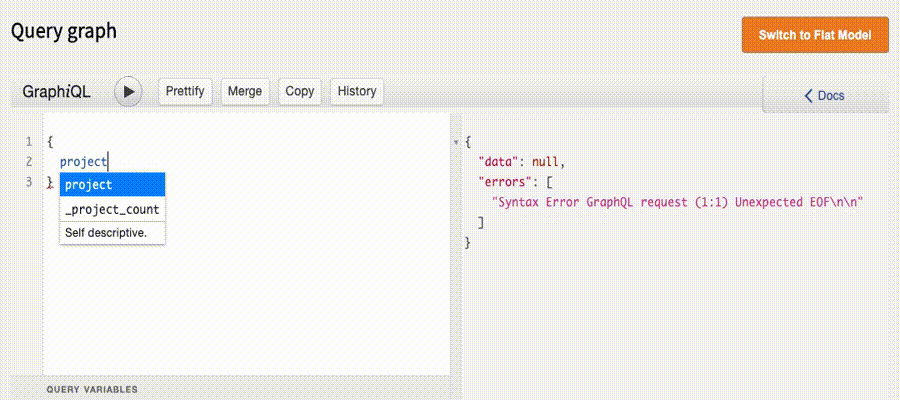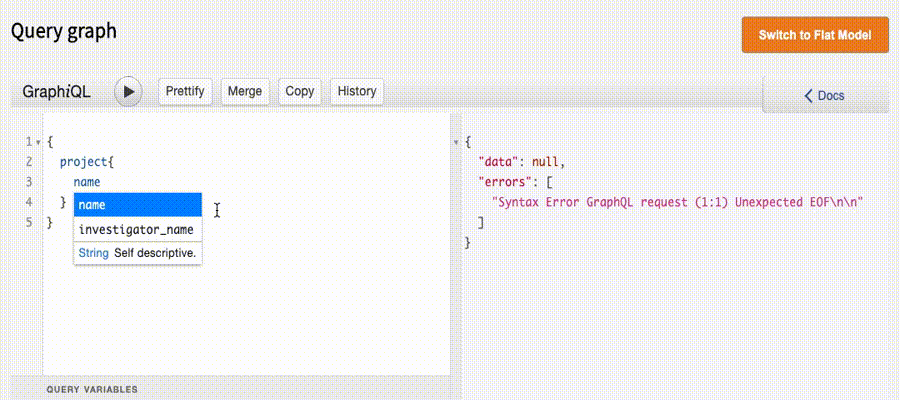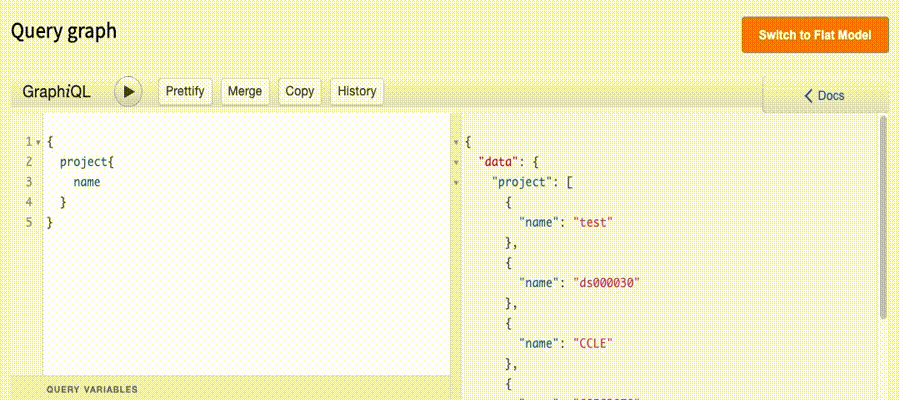
In the Graph Model, our microservice Peregrine converts GraphiQL queries and hits the PostgreSQL database.
{
_node_type (category: "medical_history") {
category
id
}
}
submitter_id: "a_submitter_id": get information for a specific submitter_idquick_search: "a_substring": get information for all files with partial matches in submitter_idfile_name: "a_filename.txt": get information for files matching a specified filename.quick_search: "sub-70080" will return files with the substring “sub-70080” in the submitter_id.file_name: "sub-70080_T1w.nii.gz" will return only files with that exact filename.submitter_id: "OpenNeuro-ds000030_sub-70080_T1w.nii_6ff0" will return only the file with that exact submitter_id, which must be unique within a node.
{
match_file_name: datanode (file_name: "sub-70080_T1w.nii.gz") {
project_id object_id id md5sum file_size file_name
data_type data_format data_category
}
match_quick_search: datanode (quick_search: "sub-70080") {
project_id object_id id md5sum file_size file_name
data_type data_format data_category
}
match_submitter_id: datanode (submitter_id: "OpenNeuro-ds000030_sub-70080_T1w.nii_6ff0") {
project_id object_id id md5sum file_size file_name
data_type data_format data_category
}
}
file_name and submitter_id arguments returns only the files that match the provided string exactly, while the quick_search argument returns all files with a submitter_id that matches the sub-string, two in this case:{
"data": {
"match_file_name": [
{
"data_category": "T1-weighted Image",
"data_format": "NII/NIfTI",
"data_type": "fMRI Image",
"file_name": "sub-70080_T1w.nii.gz",
"file_size": 11427935,
"id": "e95e513e-b76e-4de9-aef5-6b9d74e2e60f",
"md5sum": "c800fe80a333e8d3439c854dea3fdad2",
"object_id": "31525da9-7b09-48ea-966d-dd9e93786ff0",
"project_id": "OpenNeuro-ds000030"
}
],
"match_quick_search": [
{
"data_category": "T1-weighted Image",
"data_format": "NII/NIfTI",
"data_type": "fMRI Image",
"file_name": "sub-70080_T1w.nii.gz",
"file_size": 11427935,
"id": "e95e513e-b76e-4de9-aef5-6b9d74e2e60f",
"md5sum": "c800fe80a333e8d3439c854dea3fdad2",
"object_id": "31525da9-7b09-48ea-966d-dd9e93786ff0",
"project_id": "OpenNeuro-ds000030"
},
{
"data_category": "Diffusion-weighted Image",
"data_format": "NII/NIfTI",
"data_type": "fMRI Image",
"file_name": "sub-70080_dwi.nii.gz",
"file_size": 39484002,
"id": "f50e6f27-8d02-49f1-b30b-9a3d32c6075d",
"md5sum": "c37bbeb51c85471a7da4f3675c836f71",
"object_id": "94b1d6ef-5e4b-4945-bf31-bb7f06881c97",
"project_id": "OpenNeuro-ds000030"
}
],
"match_submitter_id": [
{
"data_category": "T1-weighted Image",
"data_format": "NII/NIfTI",
"data_type": "fMRI Image",
"file_name": "sub-70080_T1w.nii.gz",
"file_size": 11427935,
"id": "e95e513e-b76e-4de9-aef5-6b9d74e2e60f",
"md5sum": "c800fe80a333e8d3439c854dea3fdad2",
"object_id": "31525da9-7b09-48ea-966d-dd9e93786ff0",
"project_id": "OpenNeuro-ds000030"
}
]
}
}
In the case that too many results are returned, a timeout error might occur. In that case, use pagination to break up the query.
For example, if there are 2,550 records returned, and the GraphiQL query is timing out with (first:3000), then break the query into multiple queries with offsets:
(first:1000, offset:0) # this will return records 0-1000
(first:1000, offset:1000) # this will return records 1000-2000
(first:1000, offset:2000) # this will return records 2000-2,550
In the Flat Model, our microservice Guppy converts GraphiQL queries and hits the Elasticsearch database. Here, queries support Aggregations for string (bin counts; number of records that each key has) and numeric (summary statistics such as minimum, maximum, sum, etc) fields. For more details see the full description on our Github repositories .
Guppy allows you to query the raw data with offset, the maximum number of rows, sorting, and filters. Queries by default return the first 10 entries. To return more entries, the query call can specify a larger number such as (first: 100).
Example:
{
subject(offset: 5, first: 100, sort: [
{
gender: "asc"
},
{
_aligned_reads_files_count: "desc"
}
]) {
subject_id
gender
ethnicity
_aligned_reads_files_count
}
}
The maximum number of results returned is 10,000, which can be requested with the (first: 10000) argument. If you need to access more than that number, we suggest using the /guppy
download endpoint
.
Aggregation query is wrapped within the _aggregation keyword. In total, five aggregations are feasible at the moment:
Example for 1) Total Count Aggregation that includes a filter:
query ($filter: JSON) {
_aggregation {
subject(filter: $filter) {
_totalCount
}
}
}
Currently, Guppy uses JSON-based syntax for filters. Filters can be text/string/number-based, combined, or nested. For more examples see the full description on our
Github repositories
.
Example for a basic filter unit:
{
"filter": {
"=": {
"gender": "Female"
}
}
}
Example query including a filter:
query ($filter: JSON) {
subject (filter: $filter, first: 20) {
gender
race
ethnicity
_matched {
field
highlights
}
}
}