The Gen3 platform for creating data commons co-locates data management with analysis workspaces, apps and tools. Workspaces are highly customizable by the operators of a Gen3 data commons and offer a variety of VM images (virtual machines) pre-configured with tools for specific analysis tasks. Custom applications for executing bioinformatics workflows or exploratory analyses may be integrated in the navigation bar as well. The following documentation primarily covers exploratory data analysis in the standard Gen3 Workspace, which can be accessed by clicking the “Workspace” icon in the top navigation bar or navigating to the /workspace endpoint.
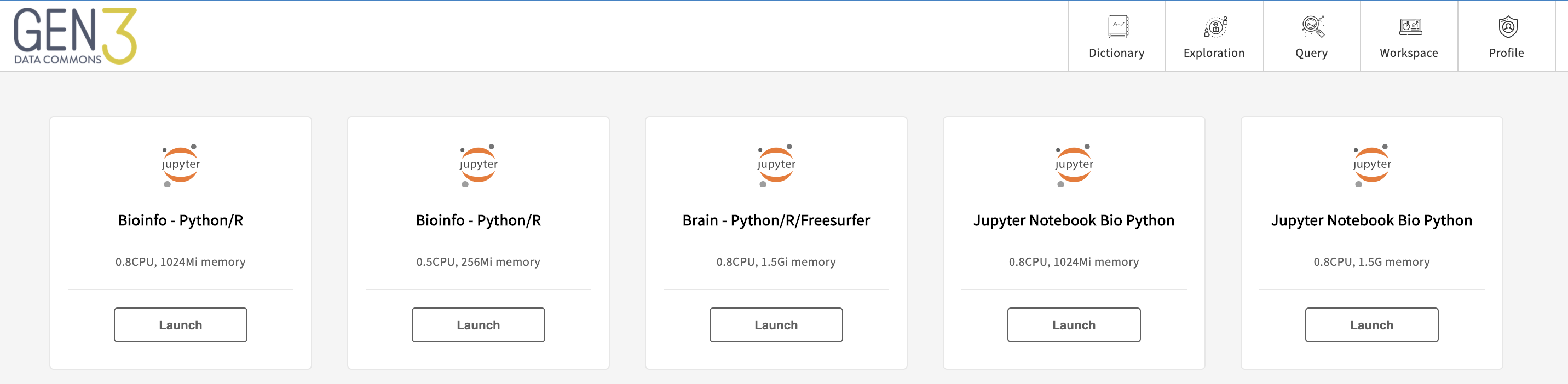
Click on the “Workspace” icon or navigate to the /workspace endpoint to see a list of pre-configured virtual machine (VM) images offered by the data commons and click the “Launch” button to spin-up a VM.
Once connected to a workspace VM, the persistent drive named “/pd” can be seen in the Files tab of either JupyterHub or RStudio. All data, notebooks, or scripts that need to persist in the cloud after workspace termination should be saved in this “pd” drive. When logging out of a workspace, this personal drive is unmounted but saved, so that when launching a new workspace VM, the drive can be mounted again to make the saved work accessible.
Jupyter notebooks and other analysis-related files like scripts can be uploaded to JupyterHub by clicking the “upload” button. Make sure to save files to the “/pd” directory if they need to remain accessible after logging out.
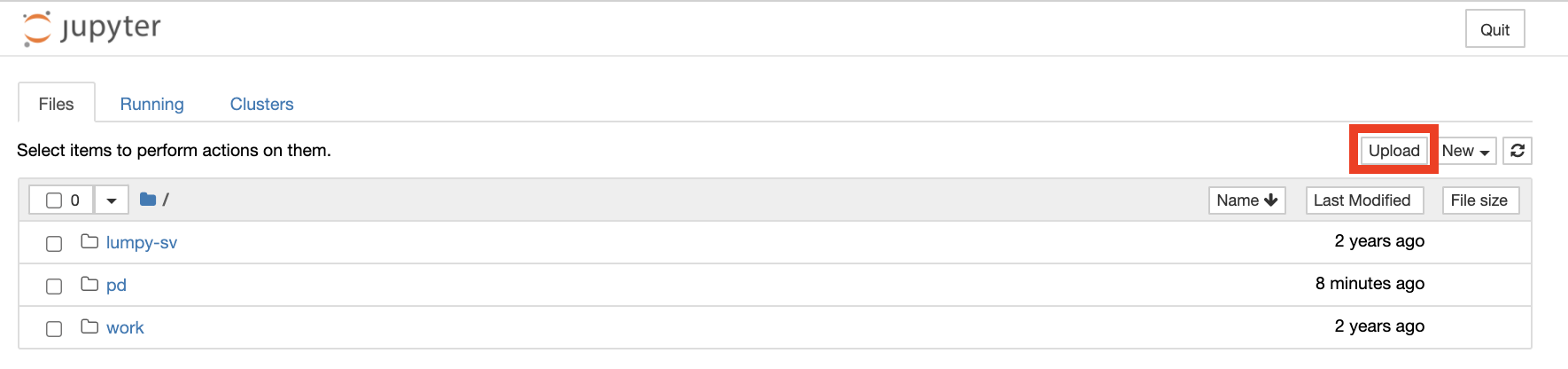
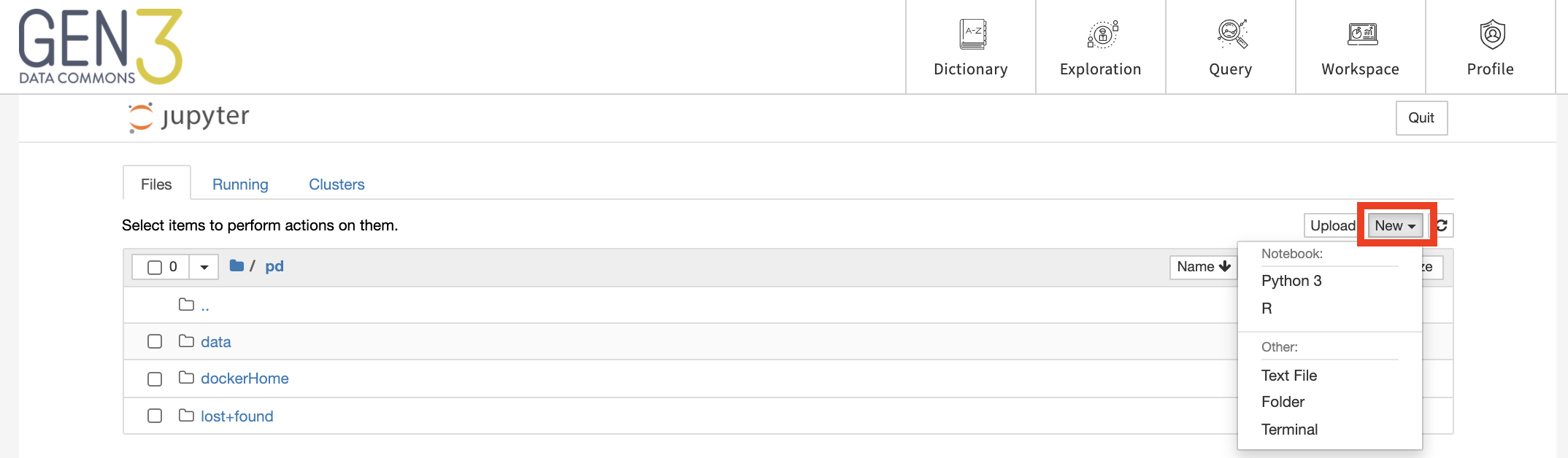
Alternatively, a new notebook can be created by clicking “New” and then choosing the type of notebook, for example, “Python 3” or “R”.
JupyterHub supports interactive programming sessions in the Python and R languages. Code blocks are entered in cells, which can be executed individually or all at once. Code documentation and comments can also be entered in cells, and the cell type can be set to support Markdown. Results, including plots, tables, and graphics, can be generated in the workspace and downloaded as files.
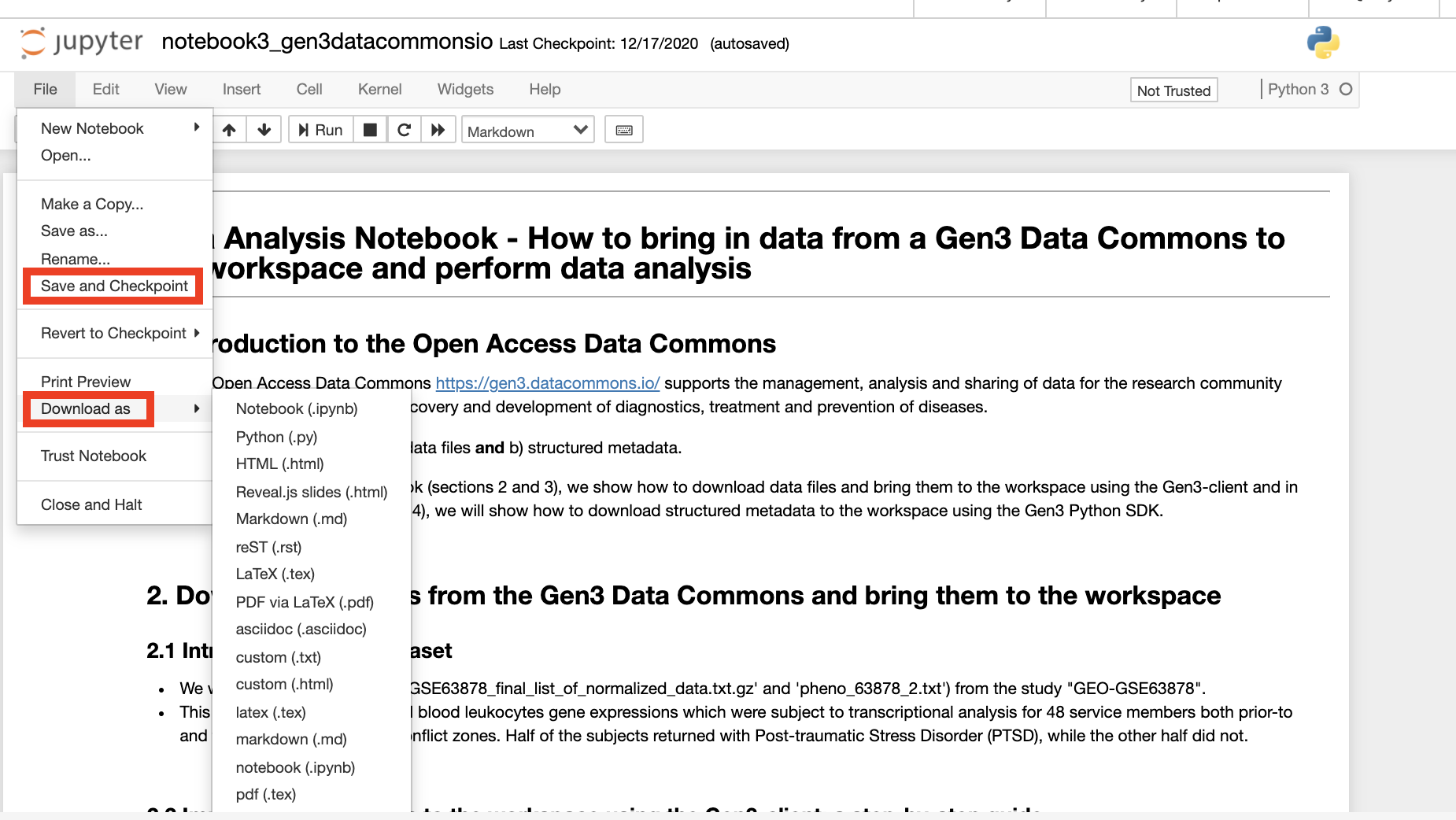
After editing a Jupyter Notebook, it can be saved in the workspace by clicking the “Save” icon or by clicking “File” and then clicking “Save and Checkpoint”. Notebooks and files can also be downloaded from the server to a local computer by clicking “File” then “Download as”.
The following clip demonstrates creating a new Jupyter Notebook in the R language.
Terminal sessions can also be started in the Workspace and used to download other tools.
You can manage active notebooks and terminal processes by clicking on the tab “Running”. Click “Shutdown” to close the terminal session or Jupyter Notebook. Be sure to save your notebooks before terminating them, and again, ensure any notebooks to be accessed later are saved in the “pd” directory.
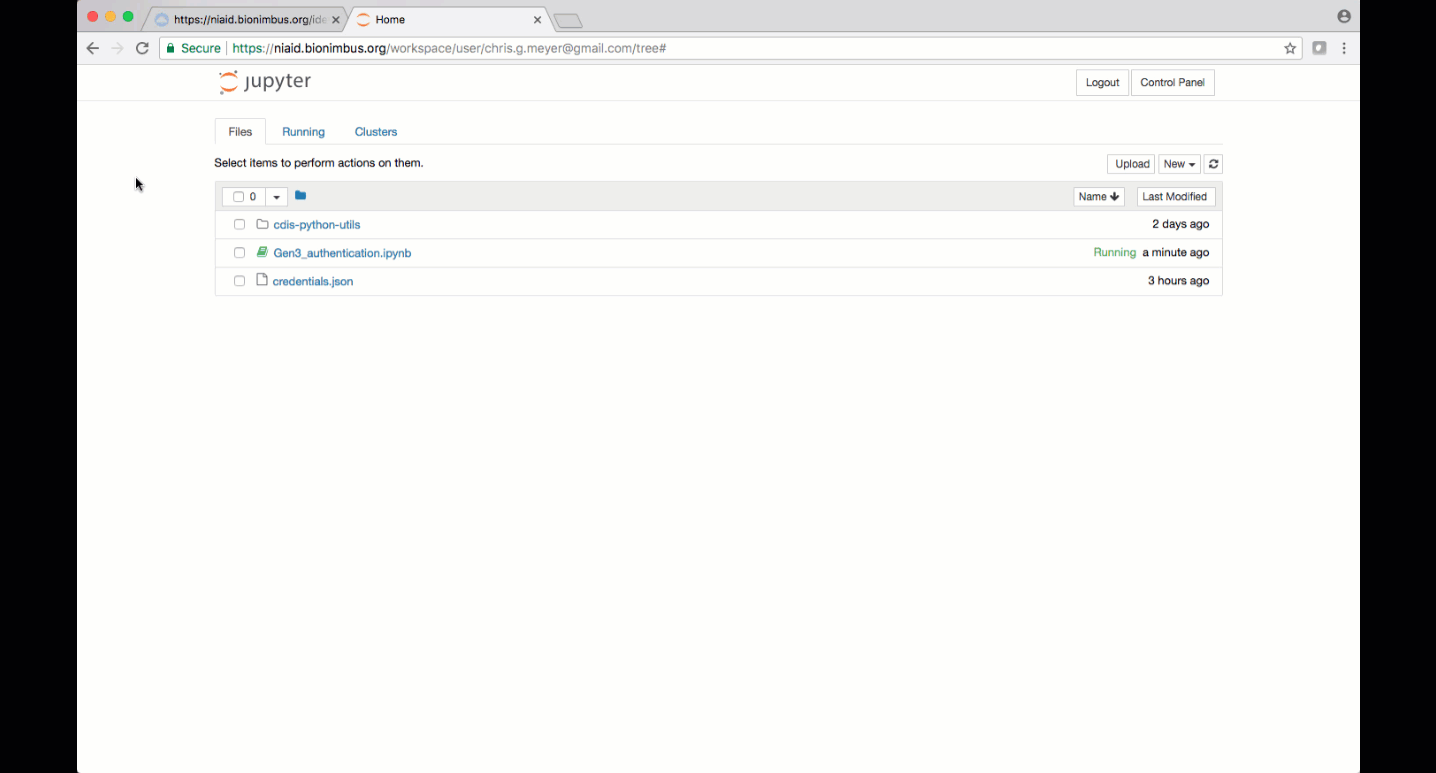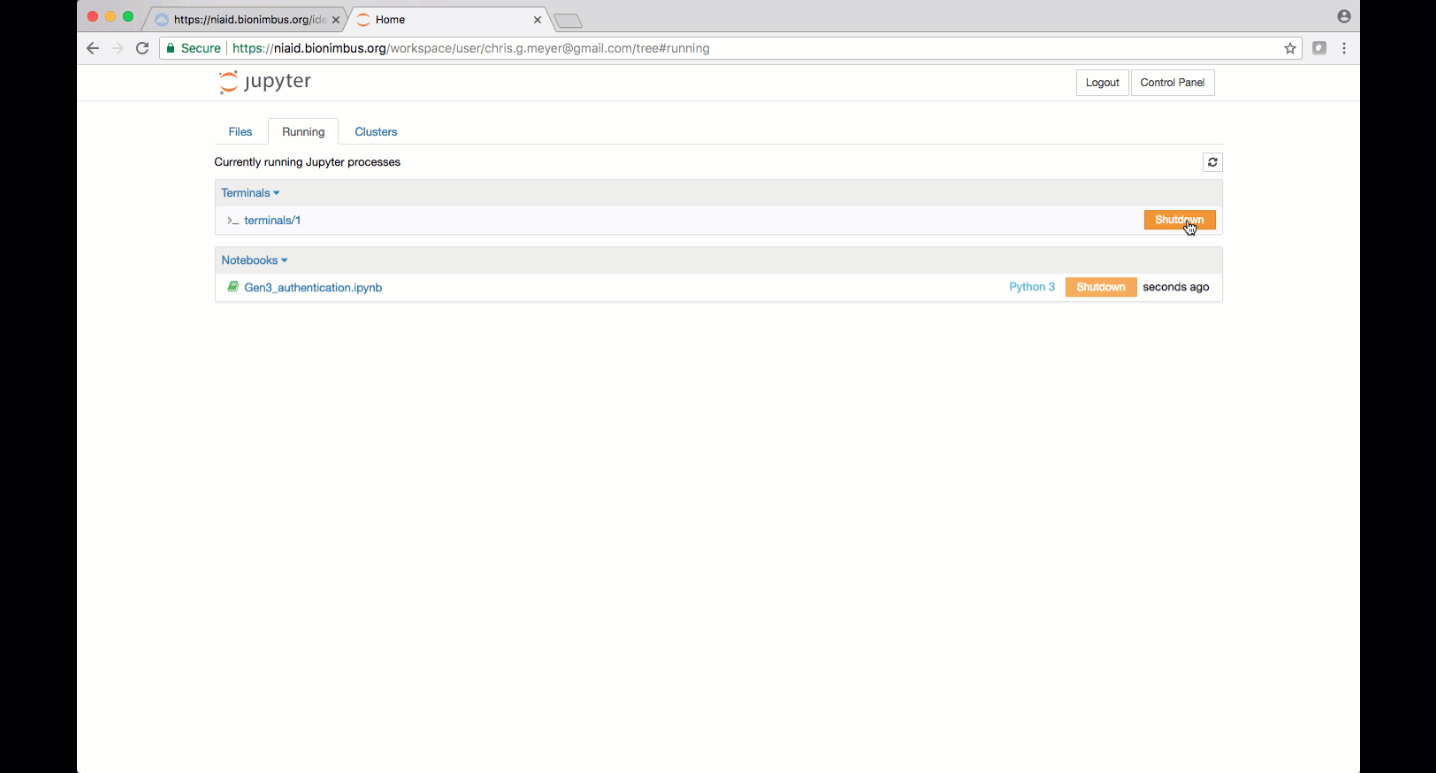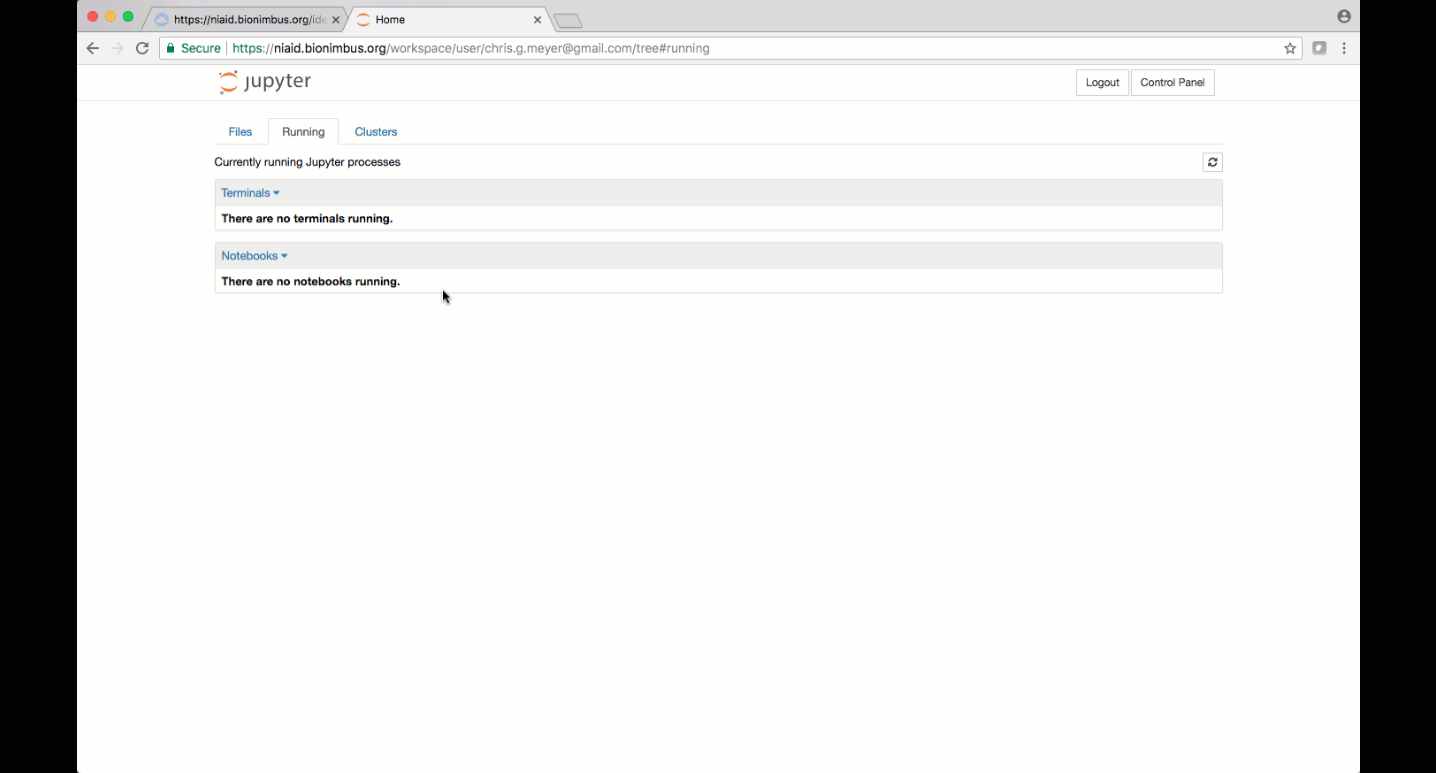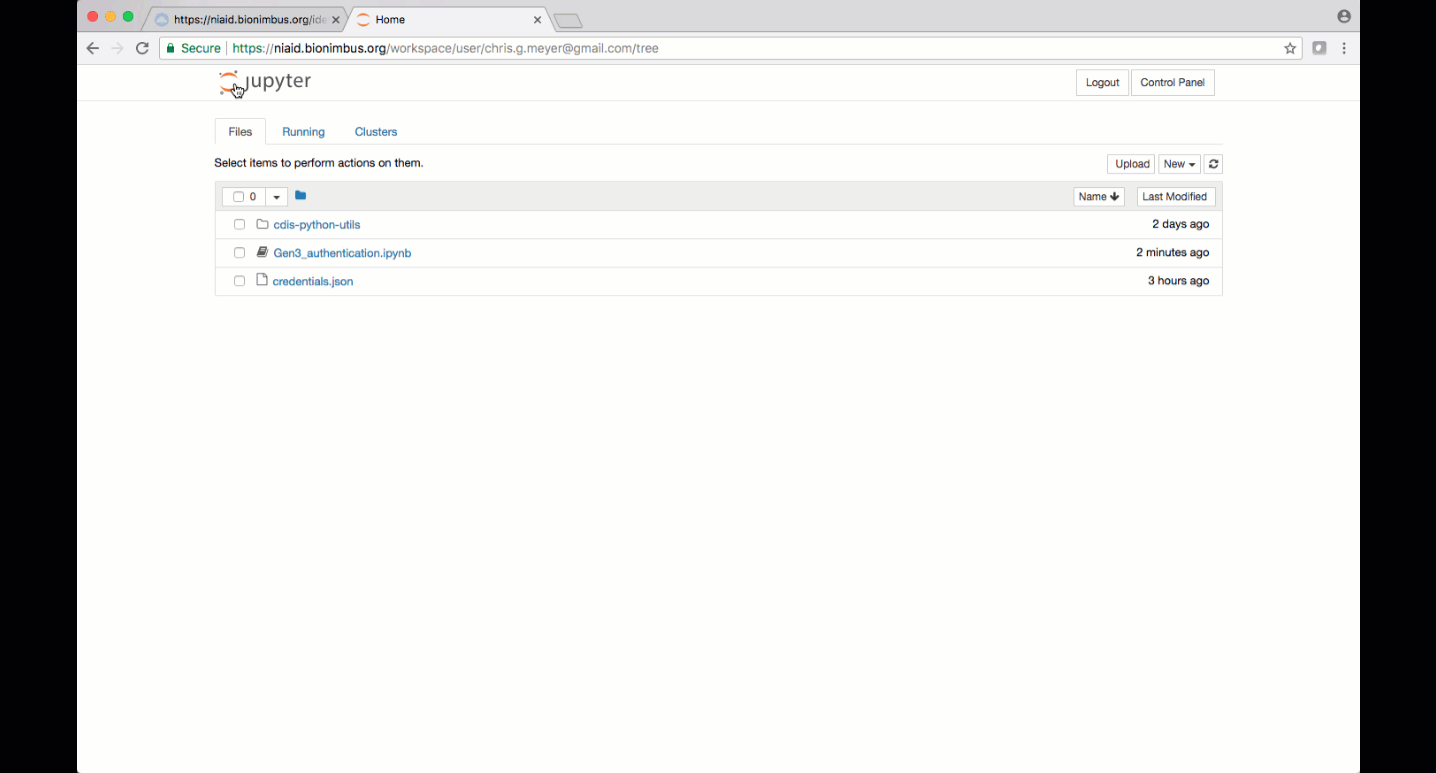
Bringing in files into the Gen3 Workspace can be achieved via the UI (directly from the Exploration page) or programmatically. Find below a description of both methods.
Note: Not every PlanX Data Commons has the function in the UI enabled and users are advised to follow available commons-specific documentation.
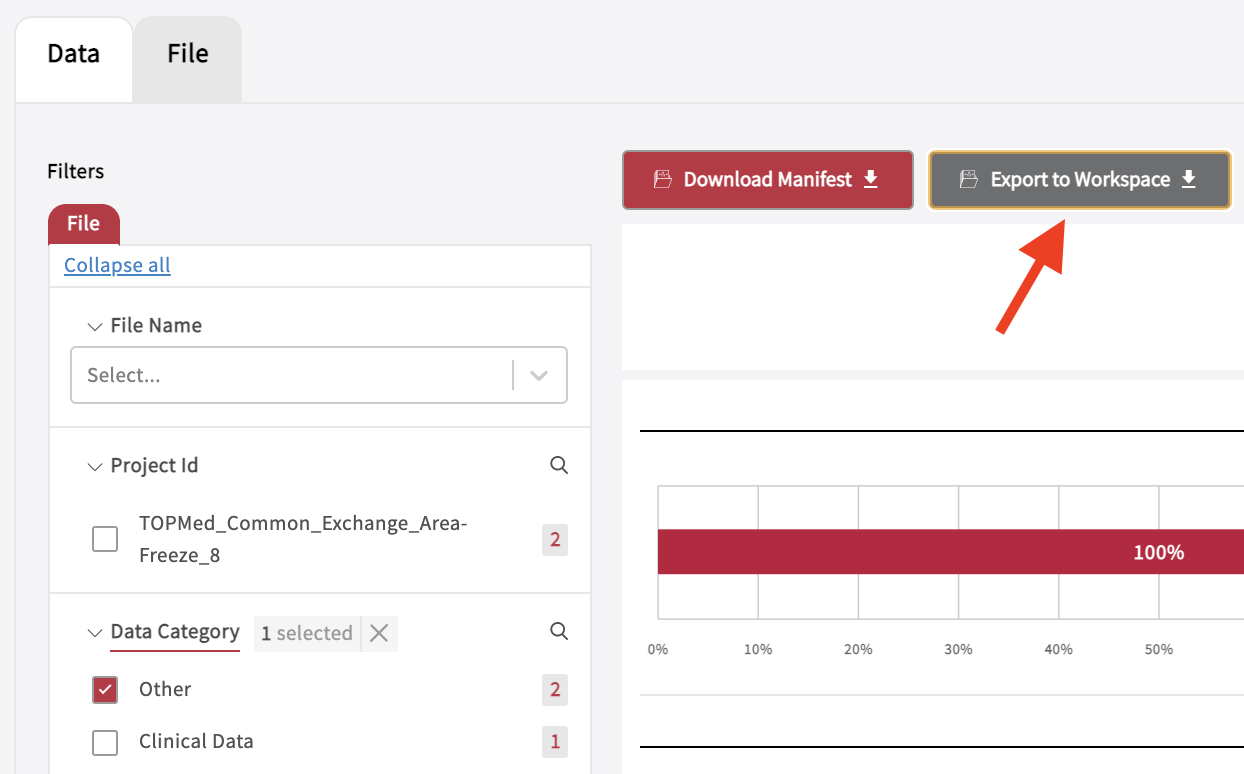
The Exploration page allows to search through data and create cohorts, which can be exported to the Workspace. After a cohort has been selected, the data can be exported to a Workspace by clicking “Export to Workspace”. Do not navigate away from the browser after clicking the button. Allow up to 5 minutes to export your files. A popup window will appear confirming that exporting a “manifest” to the workspace has been successful. Find the data or data files in the folder “data” on your persistent drive “/pd”. Please note, that the workspace mounts up to 5 different manifests while the workspace is running, but shows only the latest exported manifest in a newly launched workspace.
In order to download data files directly and programmatically from a Gen3 data commons into your workspace, install and use the gen3-client in a terminal window from your Workspace. Launch a terminal window by clicking on the “New” dropdown menu, then click on “Terminal”.
From the command line, download the latest
Linux version of the gen3-client
using the wget command. Next, unzip the archive and add it to your path:
Example:
wget https://github.com/uc-cdis/cdis-data-client/releases/download/2020.11/dataclient_linux.zip
unzip dataclient_linux.zip
PATH=$PATH:~/
Now the gen3-client should be ready to use in the JupyterHub terminal.
Other required files, like the credentials.json file which contains API keys needed to configure a profile or a download manifest.json file can be uploaded to the workspace by clicking on the “Upload” button or by dragging and dropping into the ‘Files’ tab. Text can also be pasted into a file by clicking “New”, then choosing “Text File”. Filenames can be changed by clicking the checkbox next to the file and then clicking the “Rename” button that appears.
Example:
jovyan@jupyter-user:~$ wget https://github.com/uc-cdis/cdis-data-client/releases/download/2020.11/dataclient_linux.zip
--2020-11-12 19:33:45-- Resolving github-production-release-asset-2e65be.s3.amazonaws.com
Connecting to github-production-release-asset-2e65be.s3.amazonaws.com
HTTP request sent, awaiting response... 200 OK
Length: 7445539 (7.1M) [application/octet-stream]
Saving to: ‘dataclient_linux.zip’
dataclient_linux.zip 100%[=====================================================>] 7.10M --.-KB/s in 0.05s
2020-11-12 19:33:45 (143 MB/s) - ‘dataclient_linux.zip’ saved [7445539/7445539]
jovyan@jupyter-user:~$ unzip dataclient_linux.zip
Archive: dataclient_linux.zip
inflating: gen3-client
jovyan@jupyter-user:~$ PATH=$PATH:~/
jovyan@jupyter-user:~$ gen3-client configure --apiendpoint=https://gen3.datacommons.io --profile=demo --cred=credentials.json
2020/11/12 19:43:51 Profile 'demo' has been configured successfully.
jovyan@jupyter-user:~$ gen3-client download-single --profile=demo --guid=6e312ac3-874d-4cad-b84b-474aa0209d49 --no-prompt --skip-completed
2020/11/12 20:06:43 Preparing file info for each file, please wait...
1 / 1 [===========================================================================================================] 100.00% 0s
2020/11/12 20:06:43 File info prepared successfully
pheno_63878_2.txt 78.53 KiB / 78.53 KiB [============================================================================] 100.00%
2020/11/12 20:06:44 1 files downloaded.
jovyan@jupyter-user:~$ mkdir files
jovyan@jupyter-user:~$ gen3-client download-multiple --profile=demo --manifest=file-manifest-demo.json --skip-completed
2020/11/12 22:18:13 Reading manifest...
18.17 KiB / 18.17 KiB [====================================================================] 100.00% 0s
2020/11/12 22:18:18 Total number of GUIDs: 98
2020/11/12 22:18:18 Preparing file info for each file, please wait...
98 / 98 [==================================================================================] 100.00% 3s
2020/11/12 22:18:21 File info prepared successfully
GSM1558792_Sample9_3.CEL.gz 4.04 MiB / 4.04 MiB [=============================================] 100.00%
GSM1558835_Sample31_1.CEL.gz 4.18 MiB / 4.18 MiB [============================================] 100.00%
GSM1558866_Sample46_3.CEL.gz 4.02 MiB / 4.02 MiB [============================================] 100.00%
GSM1558804_Sample15_3.CEL.gz 4.19 MiB / 4.19 MiB [============================================] 100.00%
GSM1558810_Sample18_3.CEL.gz 4.16 MiB / 4.16 MiB [============================================] 100.00%
GSM1558854_Sample40_3.CEL.gz 4.20 MiB / 4.20 MiB [====================....
2020/11/12 22:18:55 98 files downloaded.
jovyan@jupyter-user:~$ mv *.gz files
To prevent unauthorized traffic, the Gen3 VPC utilizes a proxy. If you are using one of the custom VMs setup, there is already a line in your .bashrc file to handle traffic requests.
export http_proxy=http://cloud-proxy.internal.io:3128
export https_proxy=$http_proxy
Alternatively, if you have a different service or a tool that needs to call out, you can set the proxy with each command.
https_proxy=https://cloud-proxy.internal.io:3128 aws s3 ls s3://gen3-data/ --profile <profilename>
Additionally, to aid Gen3 Commons security, the installation of tools from outside resources is managed through a whitelist. If you have problems installing a tool you need for your work, contact support@datacommons.io and with a list of any sites you might wish to install tools from. After passing a security review, these can be added to the whitelist to facilitate access.
To make programmatic interaction with Gen3 data commons easier, the bioinformatics team at the Center for Translational Data Science (CTDS) at University of Chicago has developed the Gen3 Python SDK, which is a Python library containing functions for sending standard requests to the Gen3 APIs. The code is open-source and available on GitHub along with documentation for using it .
The SDK includes the following classes of functions:
Below is a selection of commonly used functions along with notebooks demonstrating their use .
The Gen3 SDK can be installed using “pip”, the package installer for Python. For installation details, see this documentation .
# Install Gen3 SDK:
pip install gen3
# To clone and develop the source:
git clone https://github.com/uc-cdis/gen3sdk-python.git
# Use the `!` magic command to clone and develop the python SDK in the workspace:
!git clone https://github.com/uc-cdis/gen3sdk-python.git
# As the Gen3 community updates repositories, keep them up to date using:
git pull origin master
1.) Most requests sent to a Gen3 data commons API will require an authorization token to be sent in the request’s header. The SDK class Gen3Auth is used for authentication purposes, and has functions for generating these access tokens. Users do need to authenticate when using the SDK from the terminal, but do not need to authenticate once being logged in and working in the workspace of a Data Commons.
From the python shell run the following:
import gen3
from gen3.auth import Gen3Auth
endpoint = "https://gen3.datacommons.io/"
creds = "/user/directory/credentials.json"
auth = Gen3Auth(endpoint, creds)
2.) The structured data in a Gen3 data commons can be created, deleted, queried and exported using functions in the Gen3Submission class.
2.1) All available programs in the data commons will be shown with the function get_programs. The following commands:
import gen3
from gen3.submission import Gen3Submission
sub = Gen3Submission(endpoint, auth)
sub.get_programs()
will return: {'links': ['/v0/submission/OpenNeuro', '/v0/submission/GEO', '/v0/submission/OpenAccess', '/v0/submission/DEV']}
2.2) All projects under a particular program (“OpenAccess”) will be shown with the function get_projects. The following commands:
from gen3.submission import Gen3Submission
sub = Gen3Submission(endpoint, auth)
sub.get_projects("OpenAccess")
will return “CCLE” as the project under the program “OpenAccess”: {'links': ['/v0/submission/OpenAccess/CCLE']}
2.3) All structured metadata stored under one node of a project can be exported as a tsv file with the function export_node. The following commands:
from gen3.submission import Gen3Submission
sub = Gen3Submission(endpoint, auth)
program = "OpenAccess"
project = "CCLE"
node_type = "aligned_reads_file"
fileformat = "tsv"
filename = "OpenAccess_CCLE_aligned_reads_file.tsv"
sub.export_node(program, project, node_type, fileformat, filename)
will return: Output written to file: OpenAccess_CCLE_aligned_reads_file.tsv
3.) The function get_record in the class Gen3Index is used to show all metadata associated with a given id by interacting with Gen3’s Indexd service. Guids can be found on the
Exploration page
under the Files tab. The following commands:
from gen3.index import Gen3Index
ind = Gen3Index(endpoint, auth)
record1 = ind.get_record("92183610-735e-4e43-afd6-7b15c91f6d10")
print(record1)
will return: {'acl': ['*'], 'authz': ['/programs/OpenAccess/projects/CCLE'], 'baseid': 'e9bd6198-300c-40c8-97a1-82dfea8494e4', 'created_date': '2020-03-13T16:08:53.743421', 'did': '92183610-735e-4e43-afd6-7b15c91f6d10', 'file_name': None, 'form': 'object', 'hashes': {'md5': 'cbccc3cd451e09cf7f7a89a7387b716b'}, 'metadata': {}, 'rev': '13077495', 'size': 15411918474, 'updated_date': '2020-03-13T16:08:53.743427', 'uploader': None, 'urls': ['https://api.gdc.cancer.gov/data/30dc47eb-aa58-4ff7-bc96-42a57512ba97'], 'urls_metadata': {'https://api.gdc.cancer.gov/data/30dc47eb-aa58-4ff7-bc96-42a57512ba97': {}}, 'version': None}
Below are three tutorial Jupyter Notebooks that demonstrate various SDK functions that may be helpful for the analysis of data in a Gen3 workspace. You can also navigate to the notebook browser of gen3.datacommons.io ( notebook browser ) to explore the notebooks.
Find this notebook “Gen3_authentication notebook” to help guide you how to authenticate from the terminal or from the workspace (download also as .ipynb file ). Note that users do need to authenticate when using the SDK from the terminal, but do not need to authenticate once being logged in and working in the workspace of a Data Commons.
Download node files, show/select data, and plot with this notebook using data hosted on the Canine Data Commons . Users can upload this notebook as an .ipynb file to the workspace of the Canine Data Commons to start their analysis. Note, that bringing in files into the workspace as explained in this notebook can be also achieved on selected Data Commons by clicking the “Export to Workspace” button on the Exploration Page; please also note, that once files are exported from the Exploration page, users do not need to authenticate anymore in the workspace.
Download data files and metadata using the gen3-client and the Gen3 SDK, respectively, and bring them into the workspace. Run gene expression analysis and statistical analysis on the data files and metadata, respectively, and plot the outcome in different scenarios. This Jupyter Data Analysis Notebook uses data hosted on the generic Data Commons gen3.datacommons.io . Upload this notebook as an .ipynb file to the workspace of the Generic Data Commons and start your analysis. Note, that bringing in files into the workspace as explained in this notebook can be also achieved on selected Data Commons by clicking the “Export to Workspace” button on the Exploration Page; please also note, that once files are exported from the Exploration page, users do not need to authenticate anymore in the workspace.
When finished, please, shut down the workspace server by clicking the “Terminate Workspace” button.