The application programming interface (API) can be a set of code, rules, functions, and URLs that allow apps, software, servers or more generally speaking, systems, to communicate with each other. The communication between APIs consists of requests and (data) responses, usually in .JSON format.
The beauty of a Gen3 data commons is that all the functionality of the data commons website is available by sending requests to the open APIs of the data commons. Typical requests at Gen3 include querying, uploading or downloading data, which leads to communication between Gen3 microservices such as the data portal Windmill or the metadata submission service Sheepdog via open APIs.
Note: The Gen3 commons uses GraphQL as the language for querying metadata across Gen3 Data Commons. To learn the basics of writing queries in GraphQL, please visit: http://graphql.org/learn .
Gen3 features a variety of API endpoints such as /submission, /index, or /graphql, which differ in how they access the resource and contain each a subset of REST (Representational State Transfer) APIs for networked applications. REST APIs are restricted in their interactions via HTTP request methods such as GET, POST, PATCH, PUT, or DELETE. The GET request retrieves data in read-only mode, POST sends data and creates a new resource, PATCH updates/modifies a resource, PUT updates/replaces a resource, and DELETE deletes a resource. At Gen3, the GET endpoint /api/v0/submission/<program>/<project>/_dictionary will for example get the dictionary schema for entities of a project, or /api/v0/submission/graphql/getschema will get the data dictionary schema in .JSON format.
Further API specifications of the Gen3 microservices can be browsed in the developer documentation and the (REST) API Swagger documentation for each Gen3 microservice can be found in the microservice’s documentation on GitHub (e.g. Sheepdog , Peregrine ).
Sending API requests can be done in any programming language (e.g. Python, Java, R). The Center for Translational Data Science (CTDS) at University of Chicago has put together a basic SDK in Python and R to help users interact with the Gen3 APIs. Below, we list examples in how to send API requests at Gen3 using Python for demonstration purposes.
The credentials that allow access to “raw” data in the object store and ssh keys to access VMs, also allow users to programmatically query or submit data to the API. This credential is used every time an API call is made.
Each API key is valid for a month and is used to receive a temporary access token that is valid for only 30 minutes. The access token is what must be sent to the Gen3 API to access data in the commons.
For users granted data access, the API key is provided on the Profile page after clicking the “Create API key” button.
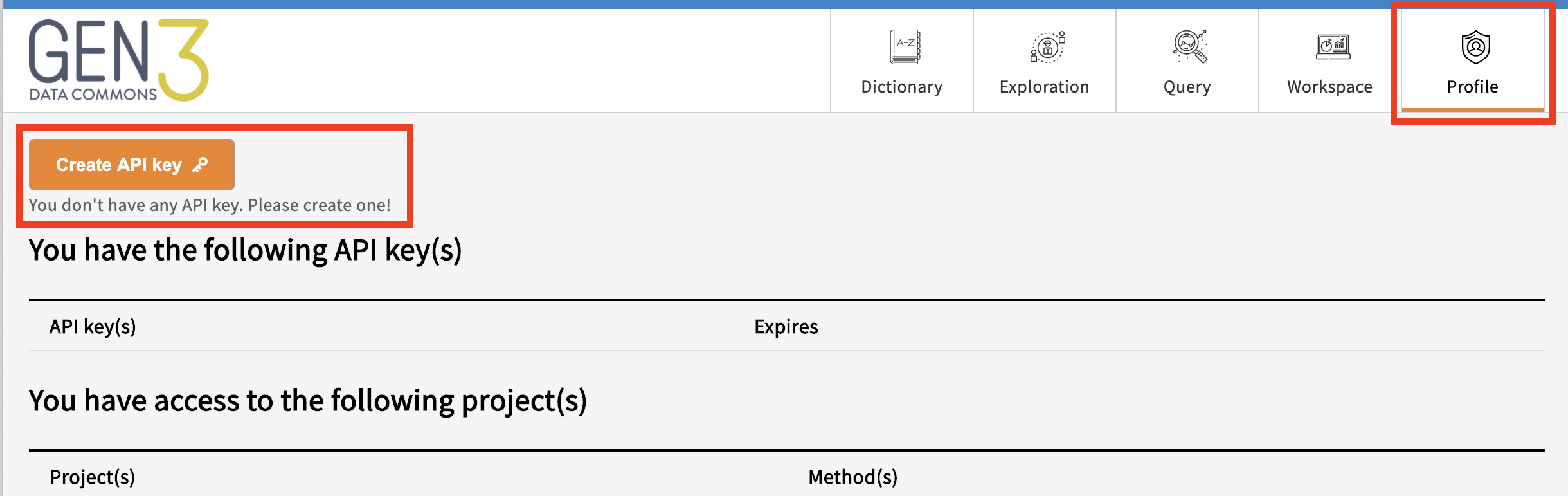
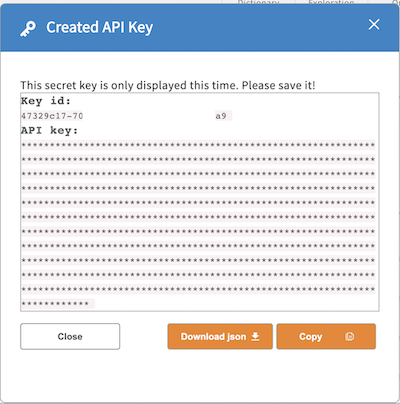
While displayed, click “copy” to copy the API key to the clipboard or “download” to download a “credentials.json” file containing the id/key pair in json format.
In Python, the following command is sent, using the module “requests”, to receive the access token:
# Save the copied credentials.json from the website and paste the api_key and key_id into a variable "key":
key = {
"api_key": "<actual-key>",
"key_id": "<a-key-uuid>"
}
# Import the "requests" Python module:
import requests
# Pass the API key to the Gen3 API using "requests.post" to receive the access token:
token = requests.post('https://gen3.datacommons.io/user/credentials/cdis/access_token', json=key).json()
# Now the access_token should be displayed when the following line is entered:
token
When submitting a graphQL query to the Gen3 API, or requesting data download/upload, include the access token in the request header:
headers = {'Authorization': 'bearer '+ token['access_token']}
# A GraphQL Endpoint Query Using the "key" JSON:
query = {'query':"""{project(first:0){project_id id}}"""};
ql = requests.post('https://gen3.datacommons.io/api/v0/submission/graphql/', json=query, headers=headers)
print(ql.text) # display the response
# Data Download via API Endpoint Request:
durl = 'https://gen3.datacommons.io/api/v0/submission/<program name>/<project code>/export?format=tsv&ids=' + ids[0:-1] # define the download url with the UUIDs of the records to download in "ids" list
dl = requests.get(durl, headers=headers)
print(dl.text) # display response
# Data Upload via API Endpoint Request:
tsv_file = '/cmd-test.tsv' # loading the tsv file that contains text string and tab-separated values
tsv = ''
count = 0
with open(tsv_file, 'r') as file:
for line in file:
if count == 0:
header = line
tsv = tsv + line + "\r"
count += 1
program_name,project_code = <program_name>, <project_code>
u = requests.put('https://gen3.datacommons.io/api/v0/submission/{}/{}'.format(program_name,project_code), data=tsv, headers=headers)
u.text # should display the API response
If an an error such as “You don’t have access… " occurs, then either you do not have access, or the API key is out of date and a new access token will need to be made. Further errors could occur if the uploaded file is not correctly formatted for the Gen3 data model.
Users with read access to a project can download individual metadata records in the project or all records in a specified node of the project using the API.
The API endpoint for downloading all the records in a single node of a project is:
{commons-url}/api/v0/submission/{program name}/{project code}/export/?node_label={node}&format={json/tsv}
Where:
{commons-url} is the gen3 data commons url (for example, 'gen3.datacommons.io'),
{program name} is the program node's property 'name' (for example, 'GEO'),
{project code} is the project node's property 'code' (for example, 'GSE63878'),
{node} is the name (or 'node_id') of the node (e.g., 'subject'),
{json/tsv} is the format in which data will be downloaded, either 'json' or 'tsv'.
For example, submitting the following API request will download all the records in the ‘sample’ node of the project ‘GEO-GSE63878’ in the demo data commons as a tab-separated values file (TSV):
https://gen3.datacommons.io/api/v0/submission/GEO/GSE63878/export/?node_label=sample&format=tsv
The API endpoint for downloading a single record in a project is as follows:
{commons-url}/api/v0/submission/{program name}/{project code}/export?ids={ids}&format={json/tsv}
Where:
{commons-url} is the gen3 data commons url (for example, 'gen3.datacommons.io'),
{program name} is the program node's property 'name' (for example, 'GEO'),
{project code} is the project node's property 'code' (for example, 'GSE63878'),
{ids} is a comma separated list of the UUIDs for the records to be downloaded (e.g., '180554c0-d0e1-41f2-b5b0-47655d7975ed,3e54268c-b4a6-4cf8-bedc-1e9e49f9d6e9'),
{json/tsv} is the format in which data will be downloaded, either 'json' or 'tsv'.
For example, submitting the following API request will download the two records corresponding to the UUIDs (the ‘id’ property) ‘180554c0-d0e1-41f2-b5b0-47655d7975ed’ and ‘3e54268c-b4a6-4cf8-bedc-1e9e49f9d6e9’, which are records in the ‘subject’ node of the project ‘GEO-GSE63878’ in the demo data commons . This downloads a ‘subjects.tsv’ file containing those two records.
https://gen3.datacommons.io/api/v0/submission/GEO/GSE63878/export?ids=180554c0-d0e1-41f2-b5b0-47655d7975ed,3e54268c-b4a6-4cf8-bedc-1e9e49f9d6e9&format=tsv
The API endpoint for querying the metadata associated with the given UUID is as follows:
{commons-url}/index/{GUID}
Where:
{commons-url} is the gen3 data commons url (for example, 'gen3.datacommons.io'),
{GUID} is the globally unique identifier (for example, 'ce214f52-1a98-4a6f-bda1-2bb2731cfd61')
The following request will show the metadata of the indexed record in .JSON format. Thus, a browser that includes a .JSON viewer (e.g. Firefox) or a manually installed plug-in can show the .JSON in pretty-print.
https://gen3.datacommons.io/index/47c46ead-f6f5-4cc9-86b9-2354cafe8c64