The following guide details the steps a data contributor must take to submit a project to a Gen3 data commons. Feel free to take a look at our webinars about data submission to our Gen3 data commons on our YouTube channel .
Data in a Gen3 data commons are either stored in variables that are exposed to the API for query (what we refer to as ‘metadata’) or are stored in files that must be downloaded prior to knowing their content (or ‘data files’). For more information on the difference between data files and metadata exposed to the API, see the documentation on the data dictionary in a Gen3 data commons .
The process of uploading a data project to a Gen3 data commons is simple:
The following sections provide step-by-step instructions for this process:
In order to upload data files, at least one record in the core_metadata_collection node must exist. If your project already has at least one record in this node, you can skip to step 2 below.
Do the following to create your first core_metadata_collection record:
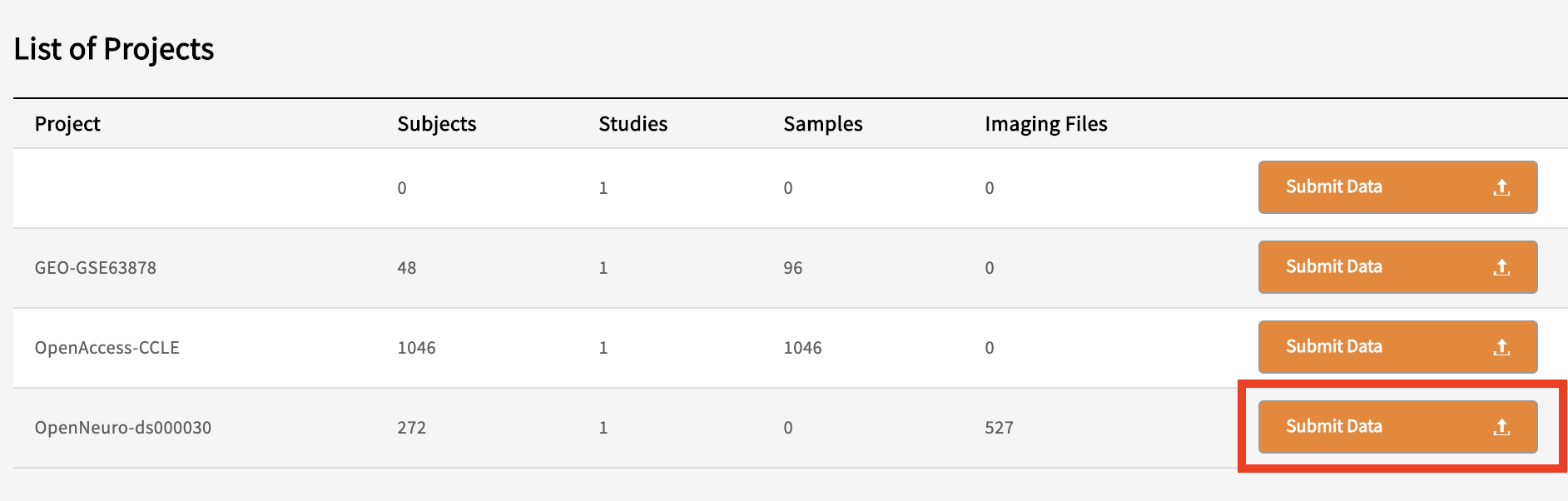
Go to your data commons’ submission portal website
Click on ‘Submit Data’
Find your project in the list of Projects and click ‘Submit Data’
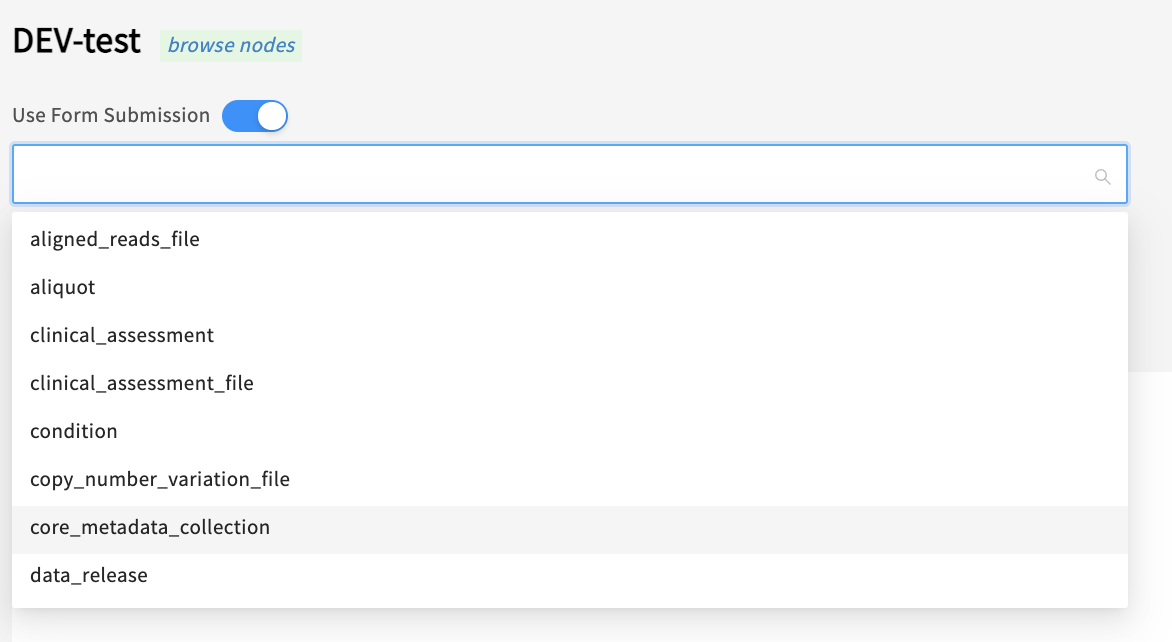
Click ‘Use Form Submission’ and choose core_metadata_collection from the dropdown list (or
edit and upload this TSV
by clicking ‘Upload File’ then ‘Submit’)
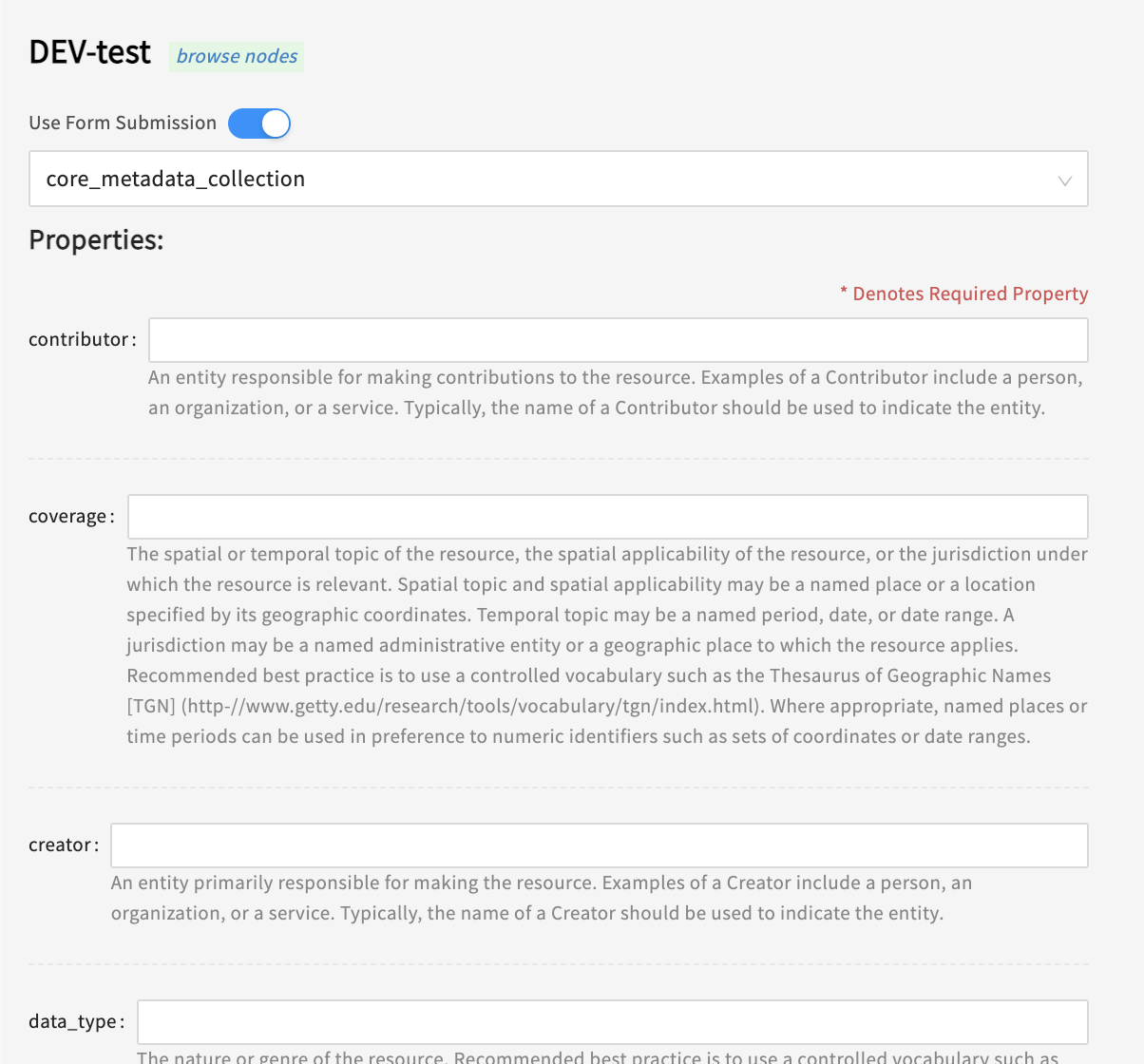
Fill in the required information (see note below)
Click ‘Upload submission json from form’ and then ‘Submit’
Make note of the submitter_id of your core_metadata_collection record for step 3 below
Note: Minimally,
submitter_idandprojects.codeare required properties. The projectcodeis the name of your project without the “program-” prefix. For example, if your project URL is https://gen3.datacommons.io/example-training , your project’scodewould be ’training’, theprogramwould be ’example’, and yourproject_idwould be the combination: ’example-training’.
You should have received the message:
succeeded: 200
Successfully created entities: 1 of core_metadata_collection
If you received any other message, then check the ‘Details’ to help determine the error.
To view the records in the core_metadata_collection node in your project, you can go to:
https://gen3.datacommons.io/example-training/search?node_type=core_metadata_collection
(replacing the first part of that URL with the URL of your actual project).
Adding files to your new Gen3 project can be done using one of two methods. The gen3-client tool (shown in steps below) offers users an easy way to upload files to Amazon s3 buckets while simultaneously indexing the files and assigning them each a unique GUID or object_id. Alternatively, if you are comfortable scripting and require something other than the default AWS bucket used by the gen3-client or your data files are already uploaded to their storage location in the cloud, we offer a command line based file submission workflow . This method offers several other benefits including the possibility of using multiple cloud resources and submitting multiple batches of data set files at once. The following documentation will focus on using the gen3-client to upload data files, including spreadsheets, sequencing data (BAM, FASTQ), assay results, images, PDFs, etc., to Amazon s3 cloud storage.
Download the latest compiled binary for your operating system.
Configure a profile with credentials downloaded from your Profile:
./gen3-client configure --profile=<profile_name> --cred=<credentials.json> --apiendpoint=<api_endpoint_url>
``
Upload Files: single data file, a directory of files, or matching files:
./gen3-client upload --profile=<profile_name> --upload-path=~/files/example.txt
./gen3-client upload --profile=<profile_name> --upload-path=~/files/
./gen3-client upload --profile=<profile_name> --upload-path=~/files/*.txt
``
For detailed instructions on configuring and using the gen3-client, visit the Gen3 client documentation .
Once data files are successfully uploaded, the files must be mapped to the appropriate node in the data model before they’re accessible to authorized users.
Go to your data commons submission portal website.
Click ‘Submit Data’.
Click ‘Map My Files’ button.
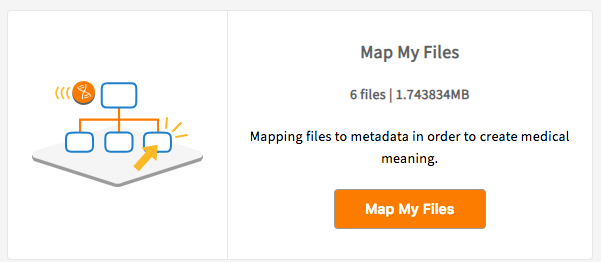
Select the files to map using the checkboxes and click ‘Map Files’ button.
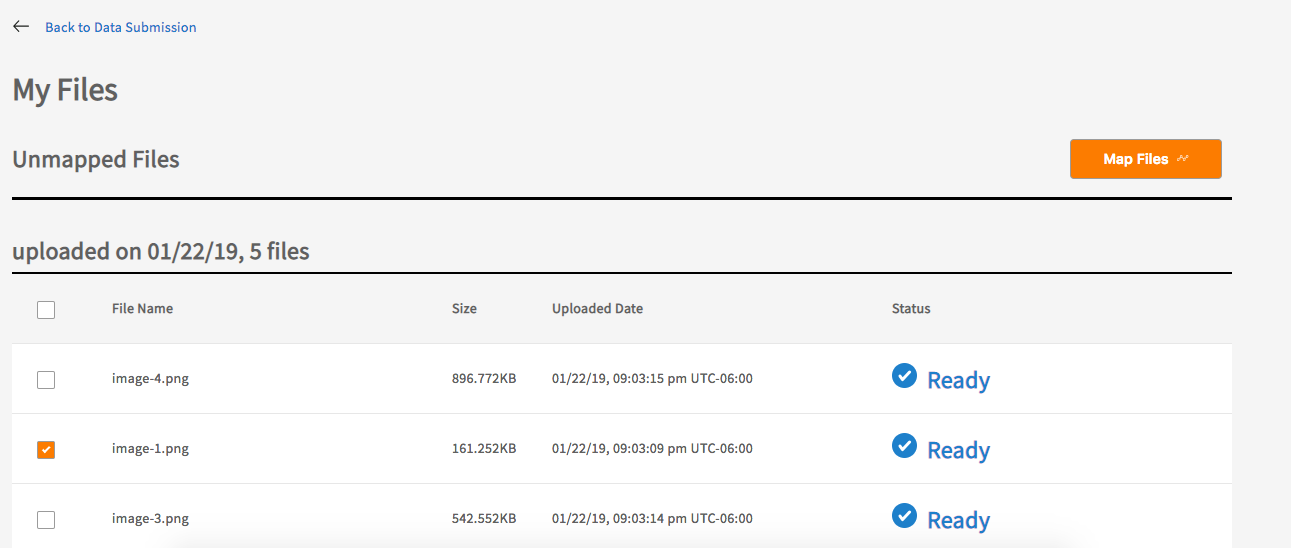
Select the project and node that the files belong to.
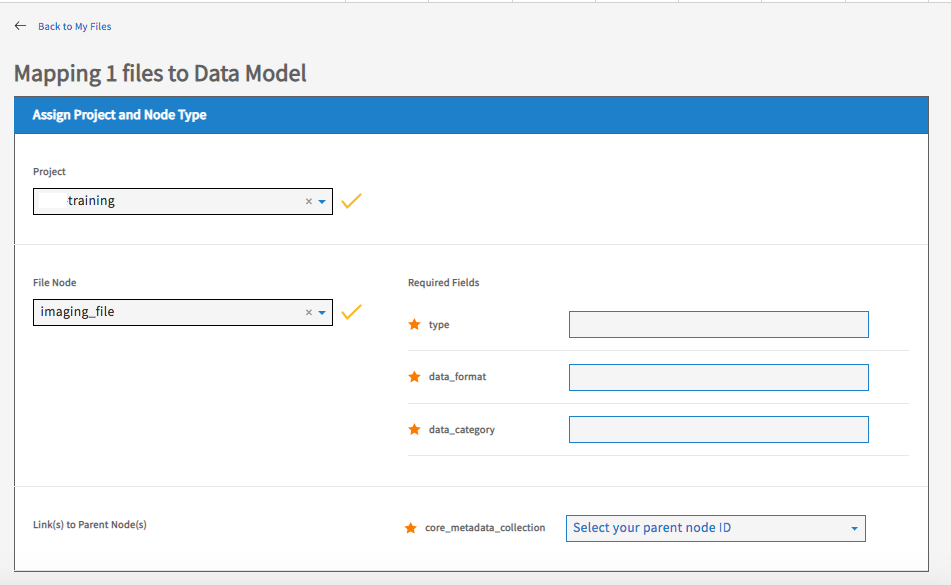
Fill in the values of any required properties and click ‘Submit’ button.
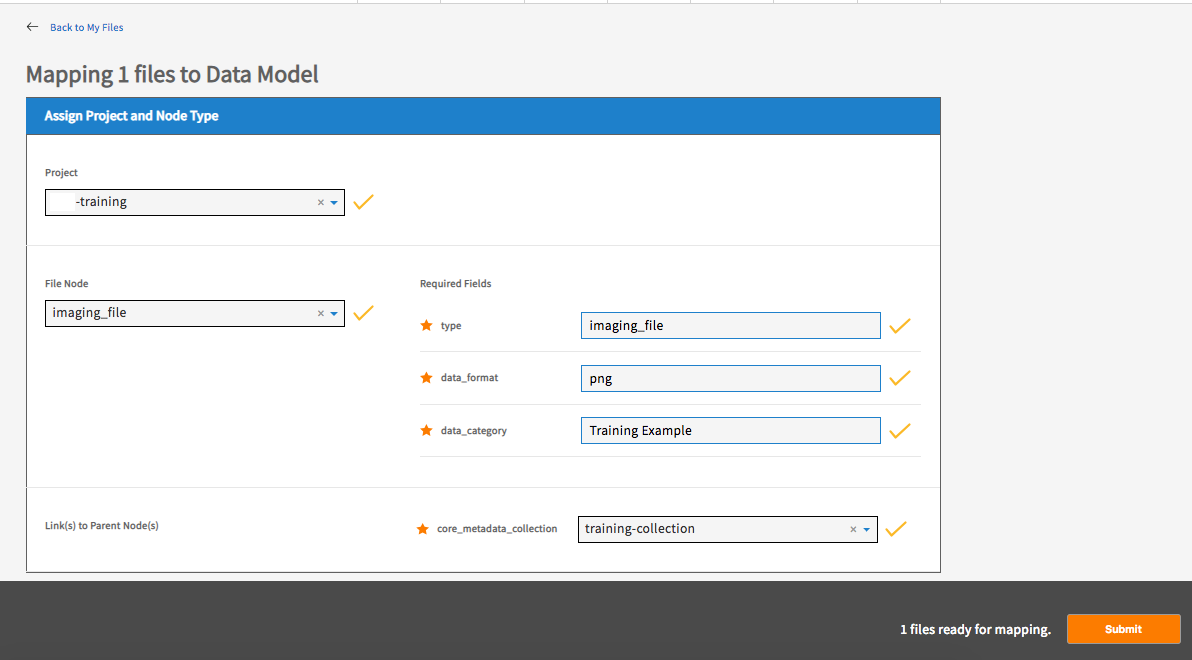
Note: The required property ‘Type’ in step 6 is the node’s name (the ’type’ of node) and should be the same as the value selected from the node dropdown list in step 5.
You should receive the message “# files mapped successfully!” upon success.
Once data files have been mapped to the appropriate data file node, the rest of the project’s metadata (e.g., patient clinical information, sample processing methods, pipeline/workflow parameters, etc.) should be submitted to the appropriate nodes. These metadata are submitted in tab-separated value (TSV) files for each node in the project, which can be downloaded from the “Dictionary” page of the data commons website.
It may be helpful to think of each TSV as a node in the graph of the data model. Column headers in the TSV are the properties stored in that node, and each row represents a “record” or “entity” in that node. When a TSV is successfully submitted, each row in that TSV becomes a single record in the node.
Properties in a node are either required or not, and this can be determined by referencing the data dictionary’s viewer’s “Required” column for a specific node.
There are a number of properties that deserve special mention:
submitter_id: Each record in every node will have a submitter_id, which is a unique alphanumeric identifier (any combination of ASCII characters) for that record across the whole project and is specified by the data submitter. It is entirely up to the data contributor what the submitter_id will be for each record in a project, but the string chosen must be unique within that project.
type: Every node has a type property, which is simply the name of the node. By providing the node name in the “type” property, the submission portal knows which node to put the data in.
id: Every record in every node in a data commons has the unique property id, which is not submitted by the data contributor but rather generated on the backend. The value of the property id is a 128-bit UUID (a unique 32 character identifier).
project_id and code: Every project record in a data commons is linked to a parent program node and has the properties project_id and a code. The property project_id is the dash-separated combination of program and the project’s code. For example, if your project was named ‘Experiment1’, and this project was part of the ‘Pilot’ program, the project’s project_id would be ‘Pilot-Experiment1’, and the project’s code would be ‘Experiment1’. Finally, just like every record in the data commons, the project has the unique property id, which is not to be confused with the project’s project_id.
Template TSVs are provided in each node’s page in the data dictionary.
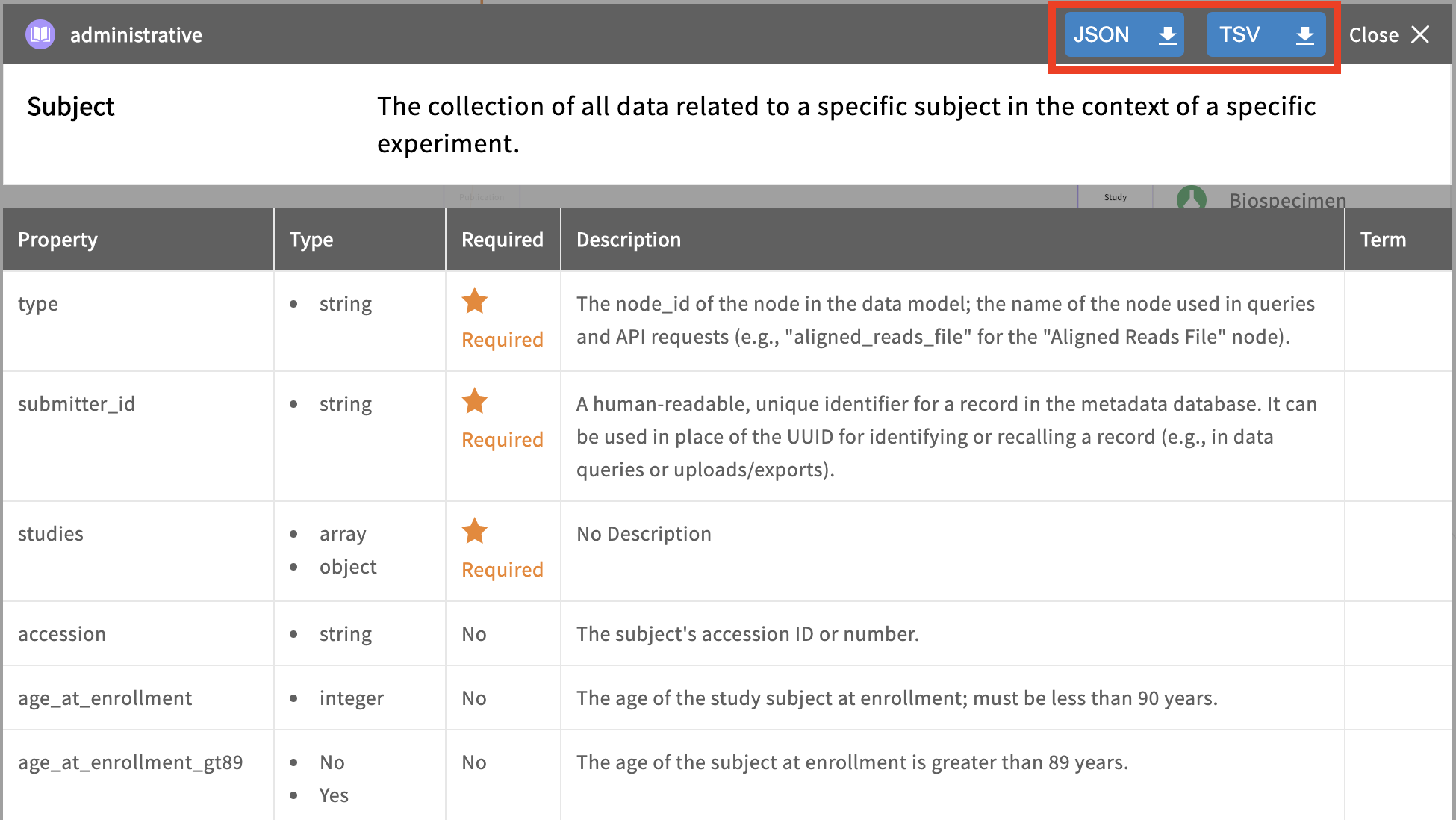
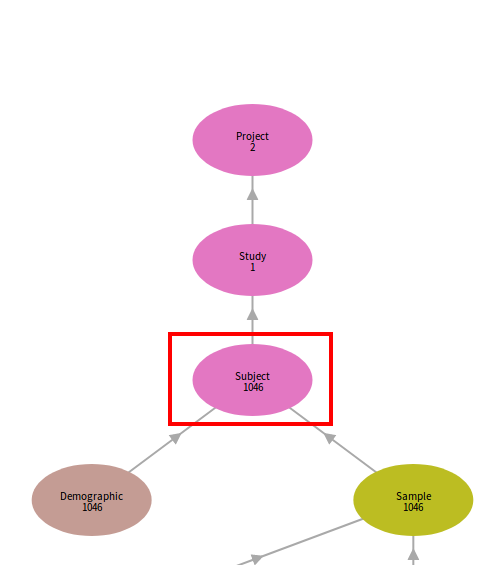
The prepared TSV files must be submitted in a specific order due to node links. Referring back to the graphical data model, a record cannot be submitted without first submitting the record(s) to which it is linked upstream (its “parent”). If metadata are submitted out of order, such as submitting a TSV with links to parent records that don’t yet exist, the validator will reject the submission on the basis that the dependency is not present with the error message, “INVALID_LINK”.
The program and project nodes are the most upstream nodes and are created by a commons administrator. The first node submitted by data contributors after core_metadata_collection depends on the specific data dictionary employed by the data commons but is usually the study or experiment node, which points directly upstream to the project node.
Often the study participants are recorded in the case or subject node, and subsequently any clinical information (demographics, diagnoses, etc.), biospecimen data (biopsy samples, extracted analytes), or other experimental methods/details are linked to each case.
At least one link is required for every record in a TSV, and sometimes multiple links could be specified. The links are specified in a TSV with the variable header <nodes>.submitter_id, where <nodes> the back-reference of the upstream node record is linking to. The value of this link variable is the specific submitter_id of the parent record. TSV or JSON templates that list all the possible link headers can be downloaded from the Data Dictionary Viewer on the data commons’ website. Properties that represent these links such as “subjects.submitter_id” or “studies.submitter_id” are array variables and can take either a single submitter_id or a comma-separated list of submitter_ids in the case that a single record links to multiple records in its parent node.
For example, there are four cases in two studies in one project. The study node was made with two study submitter_ids: “study-01” and “study-02”. The “case.tsv” file uploaded to describe the study participants enrolled will have a corresponding study.
| case | submitter_id | studies.submitter_id |
|---|---|---|
| 1 | case_1 | study-01 |
| 2 | case_2 | study-02 |
| 3 | case_3 | study-01 |
| 4 | case_4 | study-01 |
In this example cases 1, 2, and 4 all belong to “study-01”, but case 2 belongs to “study-02”. All the cases have different submitter_ids and these will be used in the subtending node that refers to a specific case.
NOTE: The
submitter_idneeds to be unique not only within one node, but across all nodes in a project. The combination ofsubmitter_idandproject_idmust be unique.
Links can be one-to-one, many-to-one, one-to-many, and many-to-many. Since a single study participant can be enrolled in multiple studies, and a single study will have multiple cases enrolled in it, this link is “many-to-many”. On the other hand, since a single study cannot be linked to multiple projects, but a single project can have many studies linked to it, the study -> project link is “many-to-one”.
Properties that represent links, like “subjects.submitter_id” or “studies.submitter_id” are array variables and can take either a single submitter_id or a comma-separated list of submitter_ids in the case that a single record links to multiple records in its parent node. Using the example above, the entry in the studies.submitter_id can be “study-01, study-02”.
In the above example, if “case_2” was enrolled in both “study-01” and “study-02”, then there would be two columns to specify these links in the case.tsv file: “studies.submitter_id#1” and “studies.submitter_id#2”. The values would be “study-01” for one of them and “study-02” for the other.
| case | submitter_id | studies.submitter_id#1 | studies.submitter_id#2 |
|---|---|---|---|
| 1 | case_1 | study-01 | |
| 2 | case_2 | study-01 | study-02 |
| 3 | case_3 | study-01 | |
| 4 | case_4 | study-01 |
To submit a TSV in the Windmill data portal:
Login to the Windmill data portal for the commons.
Click on “Submit Data” in the top navigation bar.

Click on “Submit Data” beside the project of interest to submit metadata.
Click on “Upload File”.
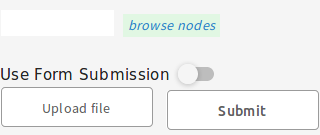
Navigate to the TSV and click “open”. The contents of the TSV should appear in the grey box below.
Click “Submit”.
A message should appear that indicates either success (green, “succeeded: 200”) or failure (grey, “failed: 400”). Further details can be reviewed by clicking on “DETAILS”, which displays the API response in JSON form. Each record/entity that was submitted, gets a true/false value for “valid” and lists “errors” if it was not valid.
For anything other than success, check the other fields for any information on the error with the submission. The most descriptive information will be found in the individual entity transaction logs. Each line in the TSV will have its own output with the following attributes:
JSON
{
"action": "update/create",
"errors": [
{
"keys": [
"species (the property name)"
],
"message": "'Homo sapien' is not one of ['Drosophila melanogaster', 'Homo sapiens', 'Mus musculus', 'Mustela putorius furo', 'Rattus rattus', 'Sus scrofa']",
"type": "ERROR"
}
],
"id": "1d4e9bb0-515d-4158-b14b-770ab5077d8b (the GUID created for this record)",
"related_cases": [],
"type": "case (the node name)",
"unique_keys": [
{
"project_id": "training (the project name)",
"submitter_id": "training-case-02 (the record/entity submitter_id)"
}
],
"valid": false,
"warnings": []
}
The “action” above can be used to identify if the node was newly created or updated. Updating a node is submitting to a node with the same submitter_id and overwriting the existing node entries. Other useful information includes the “id” for the record. This is the GUID for the record and is unique throughout the entirety of the data commons. The other “unique_key” provided is the tuple “project_id” and “submitter_id”, which is to say the “submitter_id” combined with the “project_id” is a universal identifier for this record.
To confirm that a data file is properly registered, enter the GUID of a data file record in the index API endpoint of the data commons: usually “ https://gen3.datacommons.io/index/index/GUID" , where “ https://gen3.datacommons.io ” is the URL of the Windmill data portal and GUID is the specific GUID of a registered data file. This should display a JSON response that contains the URL that was registered. If the record was not registered successfully, it is likely an error message will occur. An error that says “access denied” might also occur if the user is not logged in or the session has timed out. Note, that for these user guides, https://gen3.datacommons.io is an example URL and can be replaced with the URL from other data commons powered by Gen3.
Note: Gen3 users can also submit metadata using the Gen3 SDK, which is a Python library containing functions for sending standard requests to the Gen3 APIs. For example, the function
submit_filefrom the Gen3Submission class will submit data in a spreadsheet file containing multiple records in rows to a Gen3 Commons. The code is open-source and available on GitHub along with documentation for using it . Furthermore, this section describes steps on how to get started.
If the submission throws errors or claims the submission to be invalid, it will be the submitter’s task to fix the submission. The best first step is to go through the outputs from the individual entities, as seen in the previous section. The errors fields will give a rough description of what failed the validation check. The most common problems are simple issues such as spelling errors, mislabeled properties, or missing required fields.
When viewing a project, clicking on a node name will allow the user to view the records in that node. From here a user can download, view, or completely delete records associated with any project they have delete access to.
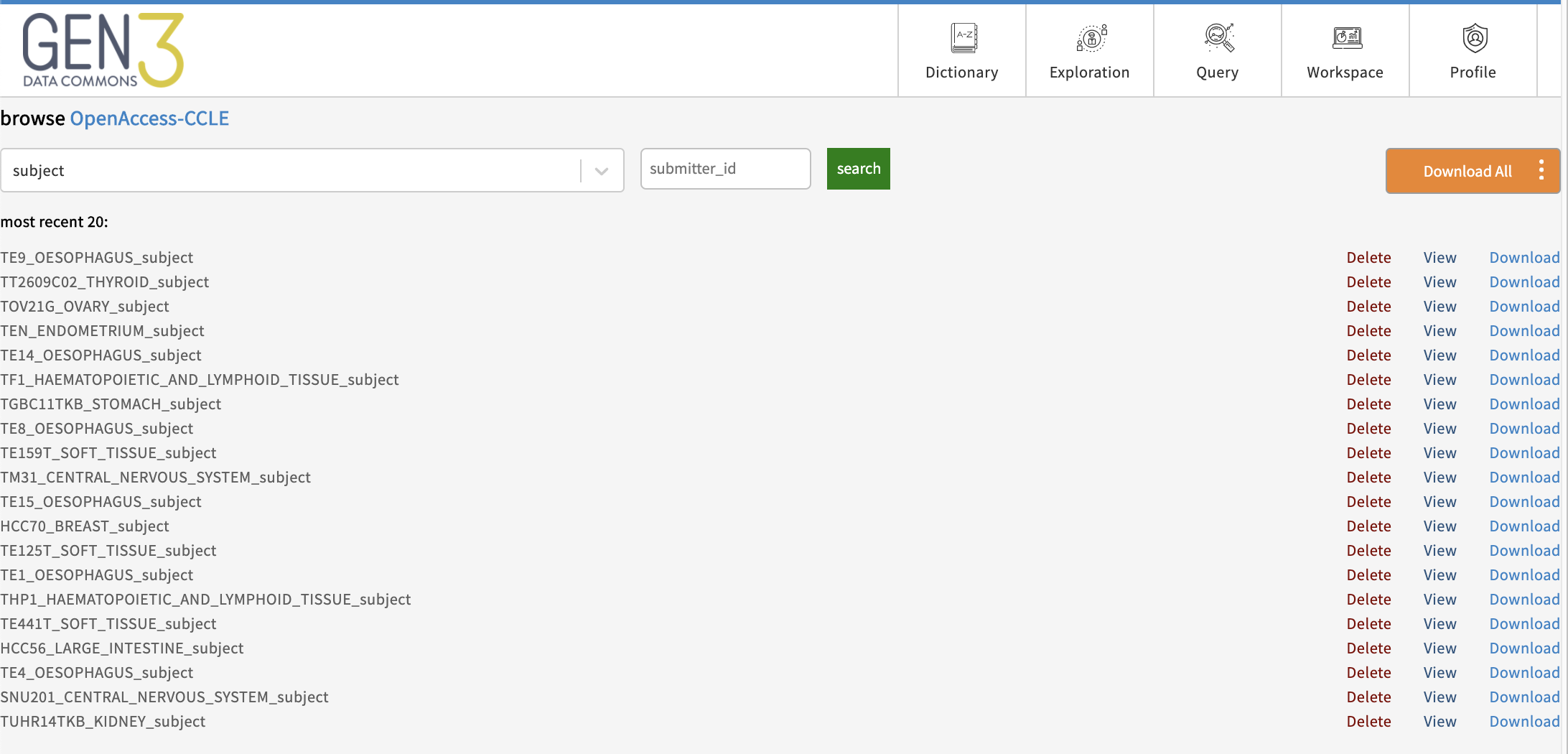
Finally, once project metadata have been submitted, data file records are linked to the corresponding records in the parent node to allow filtering or querying of submitted data files based on these experimental/clinical metadata.
The easiest way to create the link between your data files’ records and the records in their parent node is as follows:
submitter_id.Note: All records in a node can be downloaded in a single TSV file by hitting the data commons’ data export API:
<commons-url>/api/v0/submission/<program>/<project>/export/?node_label=<node>&format=tsv
For example, the following link would download a single TSV containing all the core_metadata_collection records in the project “example-training” from the commons “
https://gen3.datacommons.io
”:
The links in the downloaded TSV can be updated by filling in the submitter_ids of the corresponding parent records, saving, and re-submitting the file to the data portal website using ‘Upload File’ as done in step 4 .